Fig. 4.
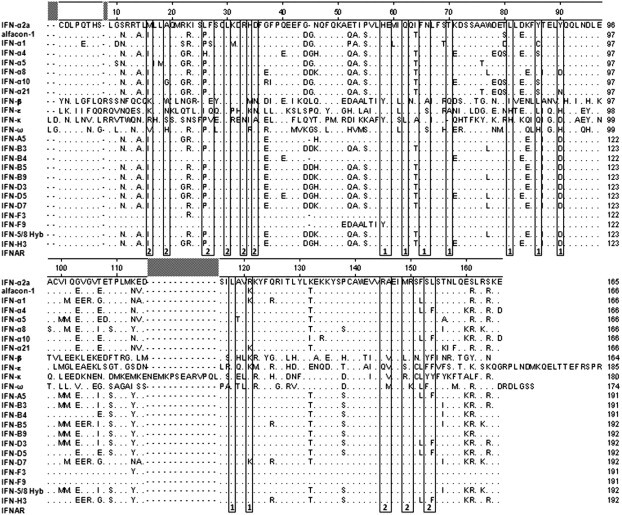
Sequence comparison of the final set of 12 hybrid IFNs to the type I IFN parents and alfacon-1. Gene sequences were determined for each of the hybrid IFNs, and derived amino acid sequences were aligned using the Clustal W method with Lasergene MegAlign (DNASTAR, Madison, WI). Numbering is according to that of IFN-alfacon-1. Amino acids identical to IFN-α2a are hidden. Residues previously shown to be important for receptor binding to IFNAR-1 or IFNAR-2 are indicated by 1 or 2, respectively, beneath the boxed amino acids (Piehler and Schreiber, 1999, Slutzki et al., 2006, Uze et al., 2007). The N-terminal extensin secretory signal peptide and the C-terminal 6x-His-KDEL sequence tags which were present on each of the hybrid IFNs are not shown in this alignment.
