Figure 1. Zika Virus (ZIKV) Infects and Kills GBM Stem Cells (GSCs) More Efficiently Than Neural Precursor Cells (NPCs).
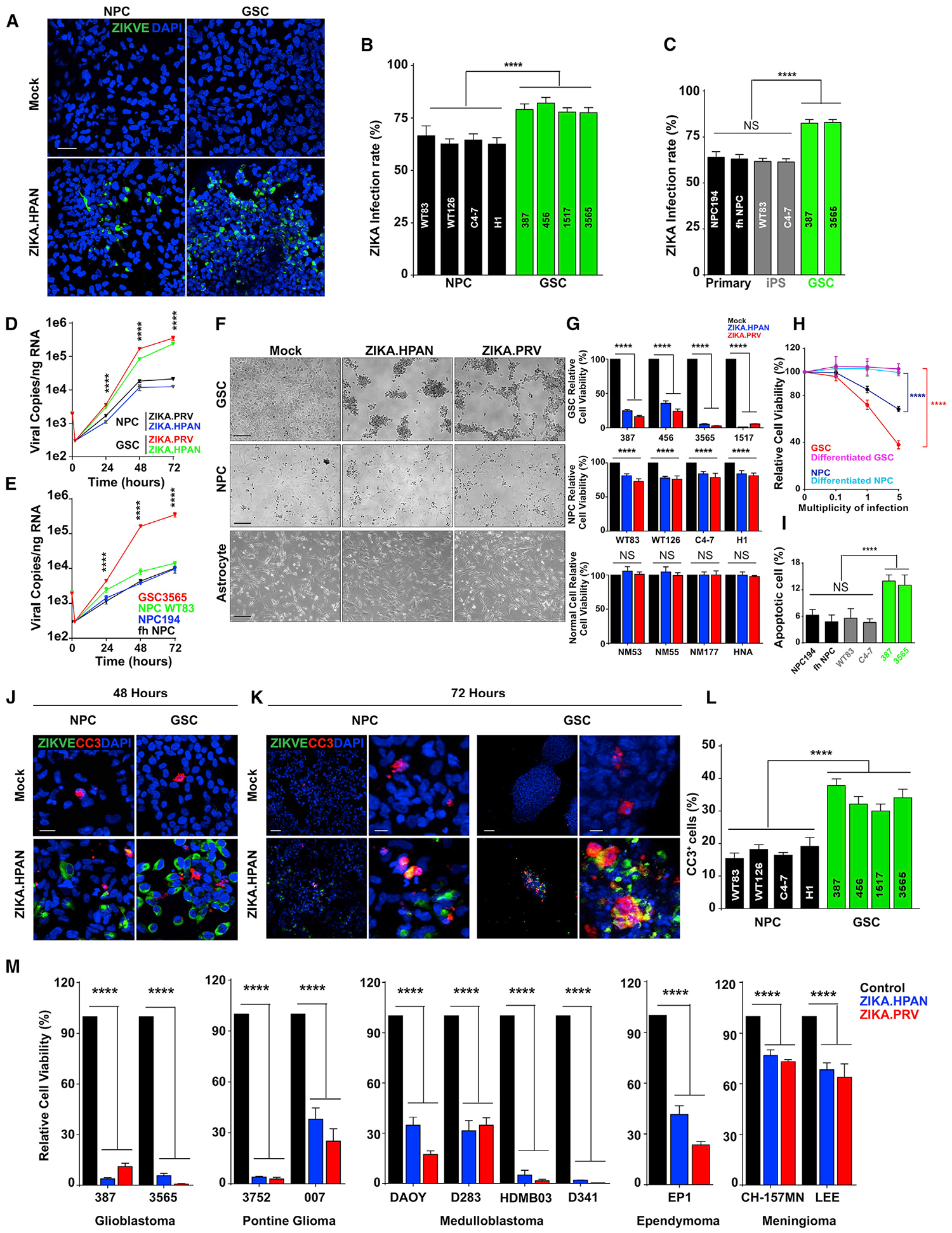
(A) Representative immunostaining for ZIKV envelope protein (ZIKV-E, green) and DAPI (blue) of GSCs and forebrain-specific NPCs 48 h post-infection (p.i.) with ZIKV. Scale bar, 50 μm.
(B) Quantification of infection efficiency in four GSC and NPC lines 48 h p.i. with ZIKV.
(C) Quantification of ZIKV+ cells in a panel of human GSCs and NPCs.
(D) Kinetics of viral RNA copies p.i. with ZIKV by measuring viral RNA copies by qRT-PCR in NPC C4–7 and GSC3565.
(E) ZIKV infection efficiency of GSCs and NPCs was measured by direct measurement of viral RNA copies.
(F) Representative bright-field images 5 days p.i. with ZIKV for GSCs, NPCs, and primary astrocytes. Scale bars, 50 μm.
(G) Cell viability normalized to day 5 mock, as measured 5 days p.i. with ZIKV for GSCs, NPCs, and primary astrocytes.
(H) GSCs (GSC3565), differentiated GSCs, NPCs (NPC C4–7), and differentiated NPCs were assayed for cell viability 72 h p.i. with ZIKV.
(I) Apoptosis of GSCs (387, 3565) and primary (NPC194, fetal human [fh] NPC) or iPSC-derived NPCs (WT83, C4–7) p.i. with ZIKV was measured by cleaved caspase-3 (CC3) staining.
(J) Representative immunostaining for ZIKV-E (green), CC3 (red), and DAPI (blue) of GSCs and forebrain-specific NPCs 48 h p.i. with ZIKV. Scale bar, 50 μm.
(K) Representative immunostaining for ZIKV-E (green), CC3 (red), and DAPI (blue) of GSCs and forebrain-specific NPCs 72 h p.i. with ZIKV. Scale bars, 50 μm.
(L) Quantification of the percentage of CC3+ cells in DAPI+ cells for GSCs and NPCs 72 h p.i. with ZIKV.
(M) Cell viability of patient-derived cultures from GBM (387 and 3565), pontine glioma (3752 and 007), meningioma (CH-157MN, IOMM-LEE), ependymoma (EP1), and medulloblastoma cell lines (DAOY, D283, HDMB03, D341) 72 h after ZIKV infection.
Experiments were performed in two biological replicates with three technical repeats. Values represent mean ± SEM. NS, no significance. ****p < 0.0001 by one-way ANOVA.
