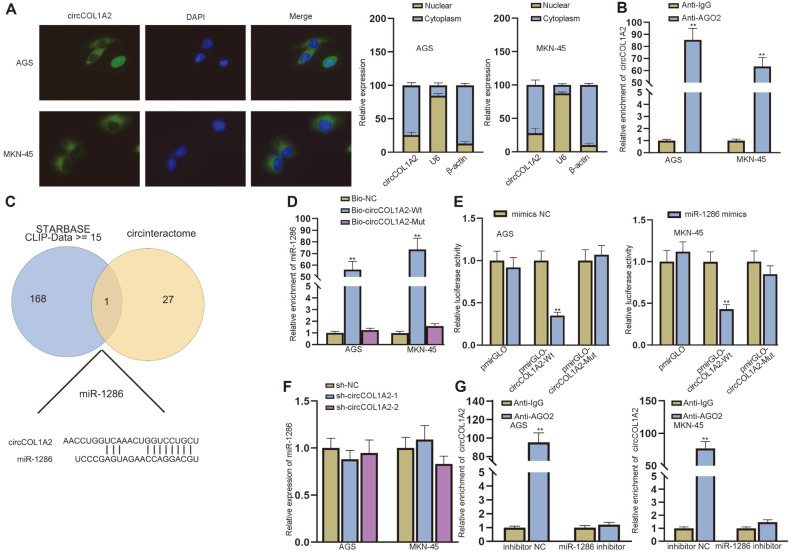
Fig. 3. CircCOL1A2 interacts with miR-1286 in GC cells.
(A) FISH and subcellular fraction assays detected the subcellular location of circCOL1A2 in AGS and MKN-45 cells. (B) RIP detected the enrichment of circCOL1A2 in Anti-AGO2 group. (C) The target miRNAs of circCOL1A2 in GC cells were predicted by starBase (http://starbase.sysu.edu.cn/) and Circular RNA Interactome (https://circinteractome.nia.nih.gov/). (D-E) RNA pull-down and luciferase reporter assays were implemented to detect the interaction between circCOL1A2 and miR-1286 in GC cells. (F) The expression of miR-1286 in GC cells was detected by qPCR after the ablation of circCOL1A2. (G) RIP detected the enrichment of circCOL1A2 in Anti-AGO2 group after the ablation of miR-1286. Student’s t-test, one-way ANOVA and two-way ANOVA were used for comparison detection. **p < 0.01.