Figure 7.
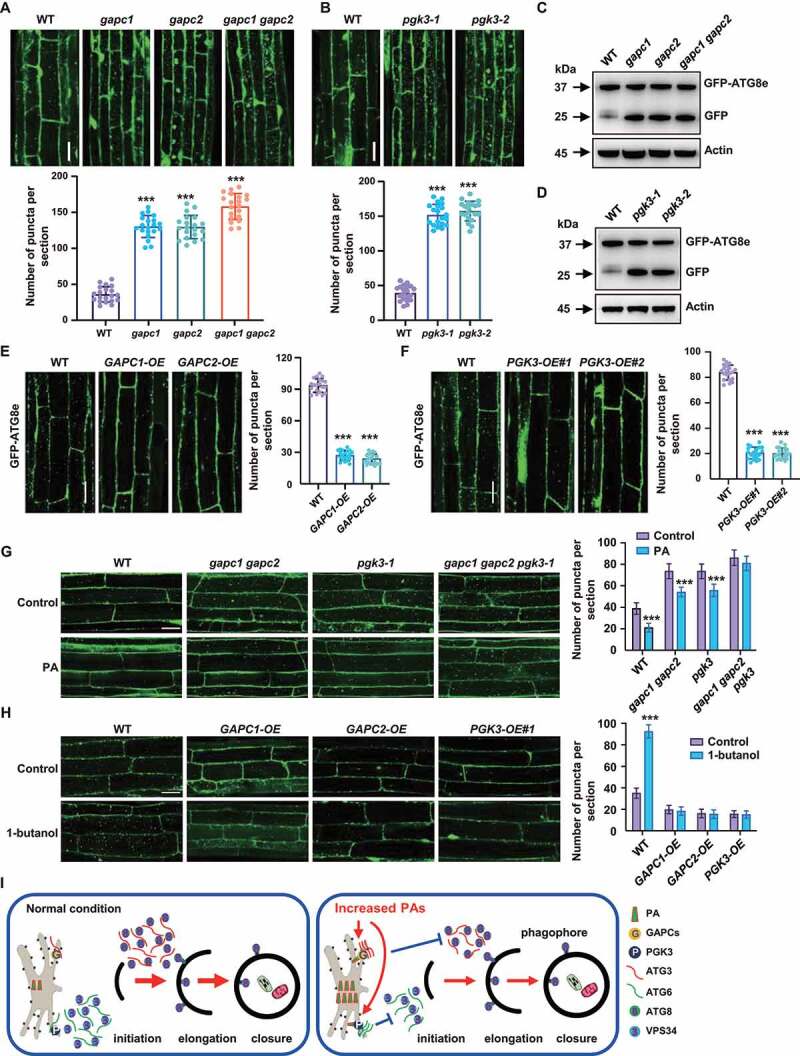
GAPCs and PGK3 inhibit autophagy in Arabidopsis. (A and B) Fluorescence observation (upper, bars: 50 μm) and quantification of puncta per root section (bottom) of WT, gapc (A) and pgk3 seedlings (B) expressing GFP-ATG8e. Five-day-old seedlings were treated with Con A (0.5 mM, 2 h) and visualized by confocal microscopy. Numbers of puncta per root section were measured and data were shown as mean ± SD (n = 20). Experiments were biologically repeated for three times. Statistical significance was determined by Student's t-test (***, P < 0.001, compared to WT). (C and D) Immunodetection of the free GFP release during autophagy-mediated vacuolar degradation of GFP-ATG8e in WT, gapc (C) or pgk3 (D) expressing GFP-ATG8e. One-week-old seedlings grown on 1/2 MS medium were transferred to 1/2 MS medium containing Con A (0.5 μM), followed by protein extraction and immunoblotting analyses. The experiments were biologically repeated for three times and ACTIN was used as a protein loading control. (E and F) Fluorescence observation (upper, bars = 50 μm) and quantification of puncta per root section (bottom) of WT, GAPCs-OE (E) and PGK3-OE (F) lines expressing GFP-ATG8e. Five-day-old seedlings were treated with Con A (0.5 mM, 4 h) and visualized by confocal microscopy. Numbers of puncta per root section were measured and data were shown as mean ± SD (n = 20). Experiments were biologically repeated for three times. Statistical significance was determined by Student's t-test (***, P < 0.001, compared to WT). (G) Observation (left) and quantitation (right) of GFP-ATG8e–labeled dots and ring-like structures in WT, gapc1 gapc2, pgk3, and gapc1 gapc2 pgk3 roots in the absence or presence of PA. Arabidopsis seedlings expressing GFP-ATG8e fusion protein were grown on 1/2 MS medium for 5 days, then transferred to 1/2 MS medium containing Con A (0.5 μM) for 8 h in the absence or presence of PA (10 μM) and visualized by confocal microscopy. Representative images were shown (bars = 50 μm) and numbers of puncta per root section were measured. Experiments were biologically repeated for three times and data were shown as mean ± SD (n = 20). Statistical significance was determined by Student's t-test (***, P < 0.001). (H) Observation (left) and quantitation (right) of GFP-ATG8e–labeled dots and ring-like structures in WT, GAPCs-OE, and PGK3-OE roots in the absence or presence of PLD inhibitor 1-butanol. Arabidopsis seedlings expressing GFP-ATG8e fusion protein were grown on 1/2 MS medium for 5 days, then transferred to 1/2 MS medium containing Con A (0.5 μM) for 8 h in the absence or presence of 1-butanol (0.4%) and visualized by confocal microscopy. Representative images were shown (bars = 50 μm) and numbers of puncta per root section were measured. Experiments were biologically repeated for three times and data were shown as mean ± SD (n = 20). Statistical significance was determined by Student's t-test (***, P < 0.001). (I) A hypothetical model illustrating how PA regulates autophagy in Arabidopsis. Under normal conditions, ATG6 activates VPS34 activity to form the PtdIns3P-enriched pre-autophagosomal structures (PAS), which organizes the site of autophagosome formation, and ATG3 catalyzes the formation of ATG8-PE conjugate to regulate the phagophore elongation. Under nitrogen or carbon starvation or other environmental stimuli, biosynthesis of phospholipids were rapidly promoted and increased phosphatidic acid (PA) enhances the interaction of glycolytic proteins GAPCs with ATG3 or PGK3 with ATG6 through direct binding GAPCs and PGK3, leading to the attenuated interactions of ATG3-ATG8e or ATG6-VPS34 thus suppressed autophagy.
