Figure 1.
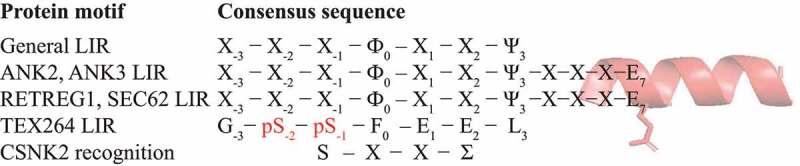
Amino acid sequence alignment of protein motifs and their upstream/downstream sequence features. The C-helix in ankyrins, ANK2 or ANK3, and in reticulophagy receptors, RETREG1 or SEC62, (red ribbon) increases binding affinity of the canonical LIR to LC3/GABARAP. A Glu residue at position 7 is often present in the C-helix. The TEX264 core LIR motif utilizes phosphorylated Ser271 and Ser272 (red) to achieve high-affinity binding to LC3/GABARAP. The TEX264 LIR fulfills the requirement for the CSNK2 recognition motif. Ф, an aromatic amino acid residue (W/Y/F); Ψ, an aliphatic residue (L/V/I); Σ, a negatively charged side chain or group (E/D/p-S/p-Y); X, any amino acid residue.
