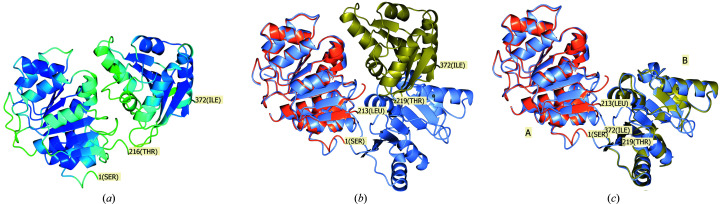
Figure 4.
(a) The unprocessed structure of human DExD-box RNA helicase DDX21, predicted by AlphaFold, coloured by the pLDDT value in the B-factor field of the PDB file. The colouring varies smoothly from blue through green to red, corresponding to pLDDT values of 100, 80 and 0, respectively. The N- and C-termini (residues 1 and 372) are labelled. Residue 216 has been labelled as a potential boundary between rigid domains found by visual inspection of the molecule. (b) The processed predicted structure superposed onto the deposited target structure, PDB entry 6l5l, in grey-blue using secondary-structure matching (SSM; Krissinel & Henrick, 2004 ▸). (c) The successful MR solution of the processed and predicted structure divided into two chains stripped of residues with pLDDT less than 70. The MR solution is superposed on the target structure with phenix.find_alt_orig_sym_mate, accounting for allowed symmetry and origin shifts. Figures were produced with CCP4mg version 2.10 (McNicholas et al., 2011 ▸).