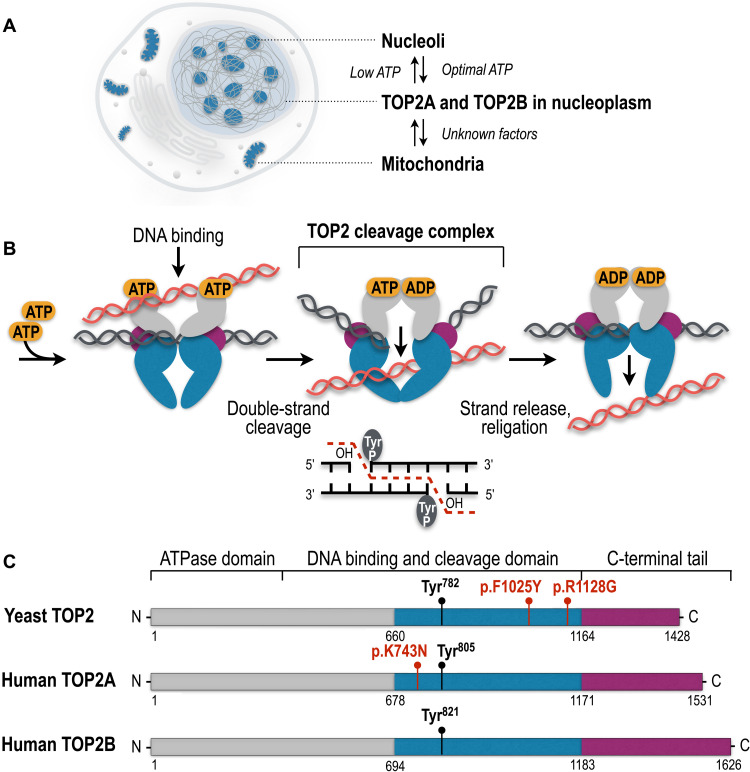
Fig. 1. Cellular localization, catalytic cycle, and evolutionary conservation of TOP2 enzymes.
(A) TOP2A and TOP2B are detected in the nucleoplasm, nucleoli, and mitochondria in the cell. (B) Eukaryotic TOP2 binds helix-helix crossovers of DNA as a homodimer that cleaves both strands of duplex DNA with 5′-four base overhangs and forms the TOP2cc by covalently attaching the catalytic tyrosine (Tyr) to the 5′-ends of the cleaved DNA. TOP2cc allows the passage of the uncleaved helix and the subsequent seamless religation of the DNA breaks. (C) Protein domains of yeast and human TOP2 enzymes. Colors show homologous regions of the enzymes. The N-terminal domain is responsible for ATP binding and hydrolysis. The central domain harbors tyrosine (Tyr) active sites to form covalent bonds with DNA and cleave and religate the DNA helix. The C-terminal domain shows the largest extent of evolutionary diversity and is involved in the association and dissociation of TOP2 with the DNA. TOP2 mutations associated with the self-poisoning phenotype and short insertions and deletions are shown in red.