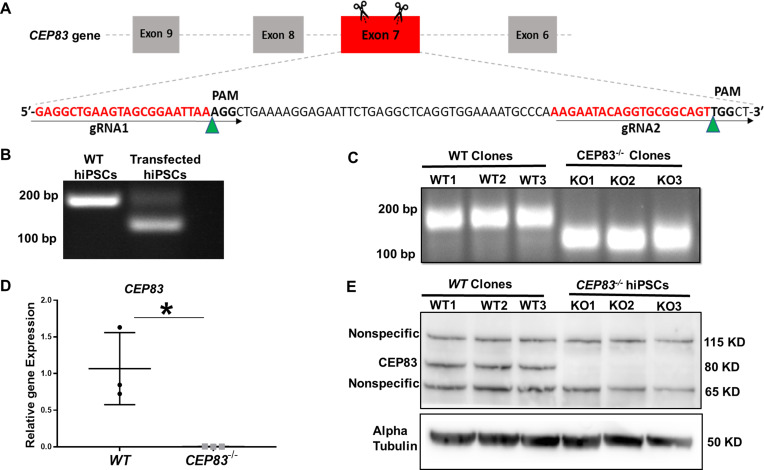
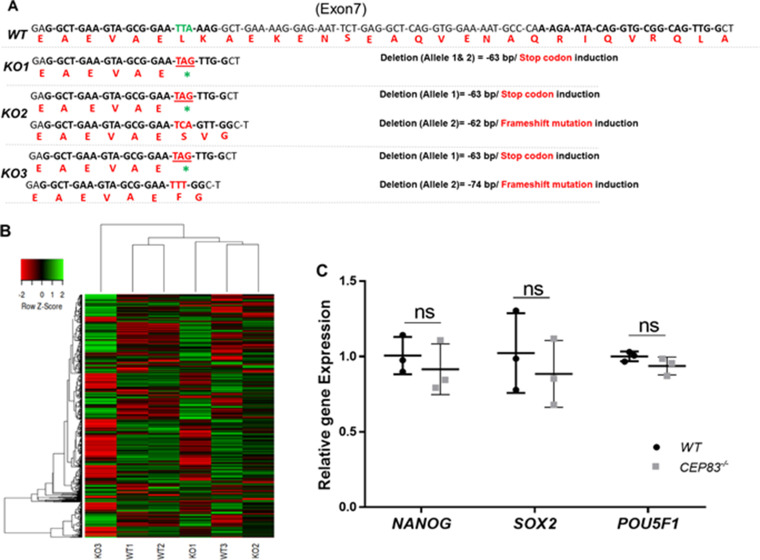
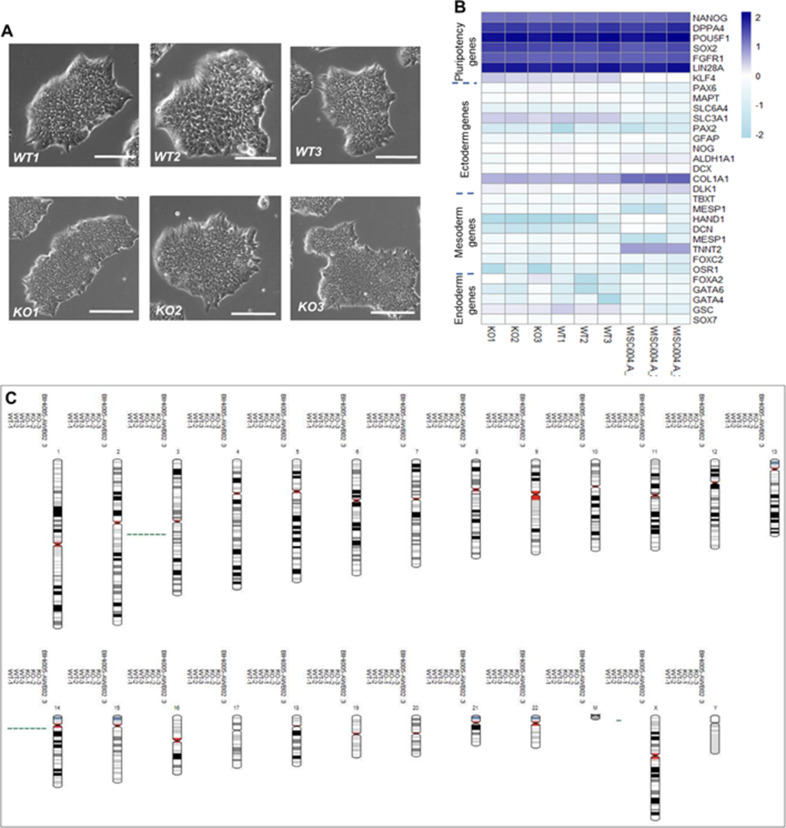
Figure 1. Generation of centrosomal protein 83 (CEP83)-deficient human pluripotent stem cells.
(A) Schematic of the experimental approach to induce a deleting mutation in exon 7 of the CEP83 gene (as described in the methods section). (B) DNA extracted from pooled transfected cells was subjected to PCR, targeting the predicted deletion site in the CEP83 gene. In addition to the 182 bp fragment present in untransfected wildtype (WT) cells, an approximately 120 bp fragment was detected in transfected cells, corresponding to the induced deletion in exon 7. (C) Three clones (CEP83−/− clones KO1, KO2, KO3) carried 62–74 bp deletions within CEP83 exon 7, which led to an induction of premature stop codons or frameshift mutation on both alleles of CEP83. Three wildtype clones (WT1, WT2, and WT3) were used as controls. (D) Quantitative RT-PCR for a fragment corresponding to the deleted region in CEP83 exon 7 produced a detectable signal in RNA extracts from WT clones but not CEP83−/− clones. (E) Immunoblotting of WT and CEP83−/− clones using a CEP83 antibody targeting the C-terminal region of the protein (see Methods for details) indicated a complete loss of the 83 KD band corresponding to CEP83 protein in the three KO clones compared with the three WT clones. Data are mean ± SD.*p<0.05 and **p<0.01 vs. WT. See Figure 1—source data 1 and 2. See also Figure 1—figure supplements 1–2.