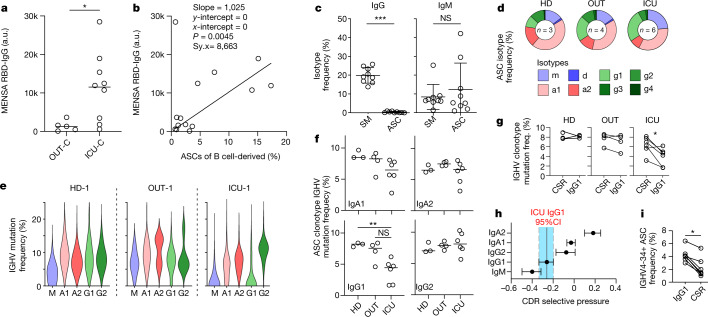
Fig. 1. Expansion of low-selection IgG1 ASC compartment is a hallmark of severe COVID-19.
a,b, MENSA samples from OUT-C (n = 7) or ICU-C (n = 9) patients were analysed for IgG reactivity against the SARS-CoV-2 RBD. RBD-specific IgG antibody in MENSA samples collected from OUT-C and ICU-C patients (a). Linear correlation of RBD-specific IgG antibody in MENSA samples versus ASC frequency of B cell-derived cells in OUT-C and ICU-C patients (b). IgG+ and IgM+ frequency of total switch memory (SM) or ASC populations from the ICU-C cohort (c). d–i, ASCs from the HD (n = 3), OUT-C (n = 4) and ICU-C (n = 6) cohorts were sorted for single B cell repertoire sequencing and subsequent analysis. Average ASC isotype compositions of HD, OUT-C and ICU-C individuals (d). Representative ASC mutation frequency distributions by isotype in HD-1, OUT-1 and ICU-1 individuals (e). IGHV gene nucleotide mutation frequencies of the indicated ASC isotypes in HD, OUT-C and ICU-C individuals (f). IGHV gene nucleotide mutation frequencies of IgG1 versus other class-switched ASCs from the indicated cohort (g). BASELINe selection analysis of CDR selection in ICU-C ASCs, grouped by isotype. Bars represent 95% confidence intervals (CI) in the group (h). IGHV4-34+ ASC frequency in IgG1 versus other class-switched ASCs (i). In a, c, g and i, statistical significance was determined using two-tailed t-test between the indicated groups. In g and i, paired analyses were used. In f, statistical significance was determined using analysis of variance with Tukey’s multiple-comparisons testing between all groups. In a–i, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. In a, c, and f, summary statistics are mean ± s.d. In h, summary statistics are mean ± 95% CI. a.u., arbitrary units; freq., frequency; NS, not significant.