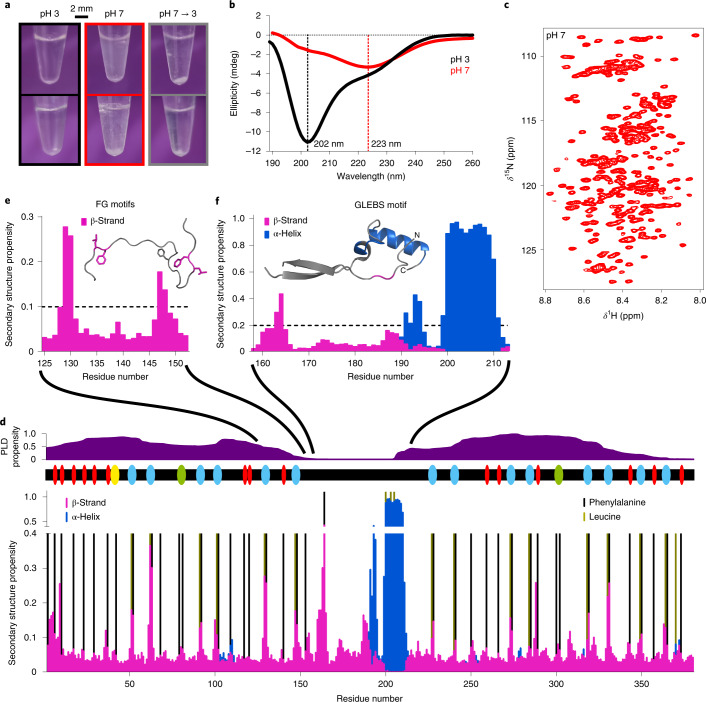
Fig. 1. Dynamic structure of Nup98FG.
a, Macroscopic changes in samples of the FG-repeat domain of Nup98FG at pH 3, after adjusting to pH 7 and then back to pH 3 before incubation (top row) and after incubation at 65 °C for 30 min (bottom row). b, CD spectra of Nup98FG in the soluble phase (pH 3) and in the condensed/aggregated phase (pH 7). c, Two-dimensional 1H–15N heteronuclear single quantum coherence spectrum of Nup98FG at pH 7 (started ~5 min after adjusting the pH from 3 to 7). d, The conformational properties of soluble Nup98FG at pH 6.8. The likelihood of residue-specific backbone torsion was determined from the experimental NMR chemical shifts using TALOS-N. The propensity for prion-like domain (PLD) structure and the location of FG motifs are shown above (red, FG; yellow, SAFG; cyan, GLFG; green, FXFG). e, The β-strand motifs in the N-terminal prion-like domain (taken from d). The conformation derived from TALOS-N is shown as the inset; the colouring is based on the threshold propensity shown in the graph. Phenylalanine and leucine side chains are displayed. f, Preformed secondary structure of the GLEBS-binding motif of monomeric unbound Nup98FG (taken from d). The inset shows the crystal structure (Protein Data Bank identification (PDB ID): 3MMY) of the GLEBS-binding motif in the complex with the mRNA export factor Rae1. Regions of the crystal structure that are preformed (>0.2) prior to binding to Rae1 are coloured (the α-helical structure is shown in blue and the β-structure in magenta).