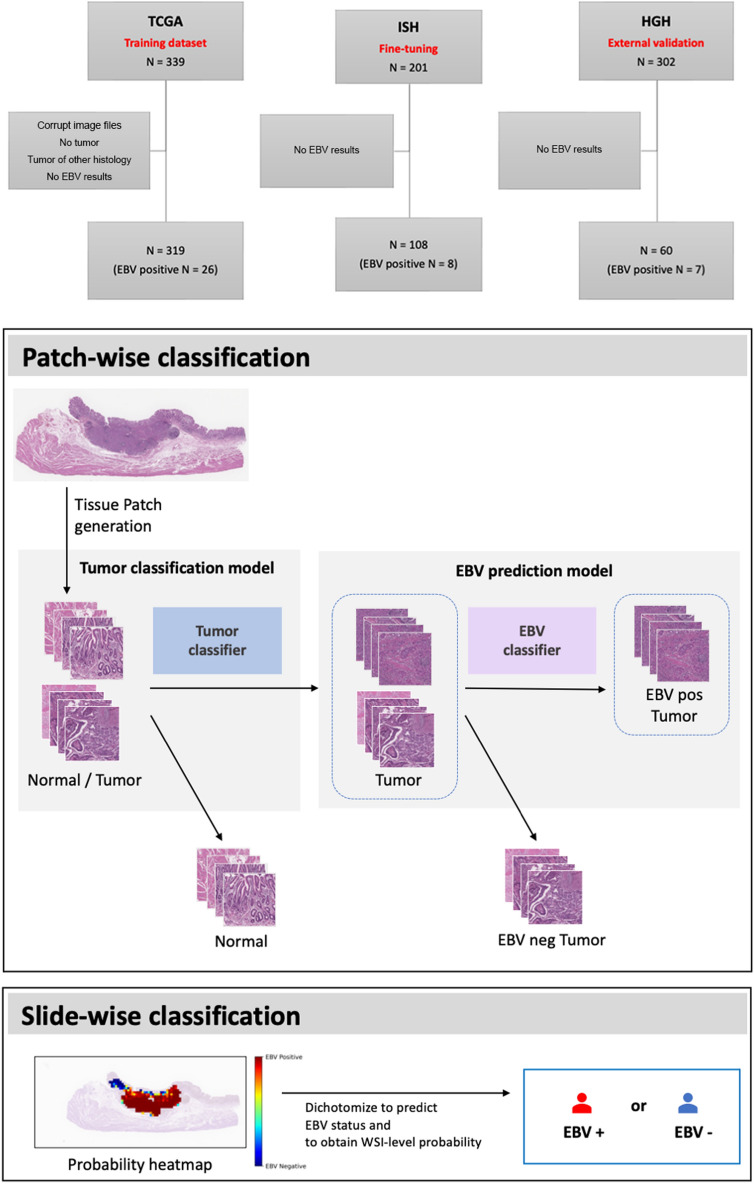
Figure 1.
Overview of EBVNet. EBVNet was trained on the TCGA dataset, and fine-tuned on the ISH dataset. EBVNet before and after fine-tuning were externally validated on the HGH dataset. Each H&E histology slide was preprocessed by discarding non tissue-containing background using Otsu’s thresholding and generating non-overlapping patches. All the patches were stain normalized (not shown in the figure). First, the tumor classification model (blue box) receives all the tissue-containing patches and returns the probability of each patch being a tumor patch. The patches with probabilities of higher than 0.5 are assigned as tumor patches, otherwise, they are assigned as normal patches. The tumor-predicted patches are then fed into the second classifier (EBV prediction model, purple box), with the resulting probability of being an EBV positive tumor patch. The tumor-predicted patches with probabilities higher than 0.1 are assigned as EBV positive tumor patches, otherwise, they are assigned as EBV negative patches. Based on the patch-wise classification result, the EBV probability score (EPS), which is the ratio of the number of EBV-positive tumor-predicted patches to the number of tumor-predicted patches, is calculated for each slide. The slide with calculated EPS higher than 0.2 is assigned as an EBV positive slide, otherwise, it is assigned as an EBV negative slide.