Fig. 1. Overview of the study.
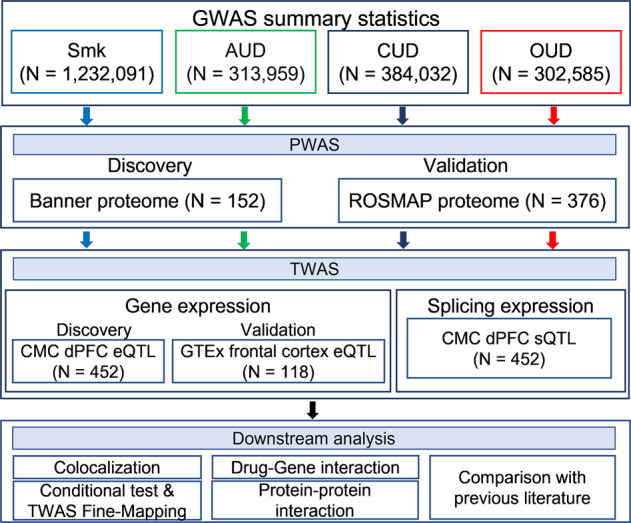
GWAS summary statistics included in the study were based on 4 substance use traits (SUT): smoking initiation (Smk), alcohol use disorder (AUD), cannabis use disorder (CUD) and opioid use disorder (OUD). For PWAS, human brain proteomes from Banner (discovery) and ROSMAP (validation) datasets were integrated with each set of GWAS summary statistics. TWAS based on brain eQTL datasets from discovery (CommonMind Consortium - CMC) and validation (Genotype-Tissue Expression - GTEx) datasets was conducted for each SUT. TWAS splicing expression analysis (CMC - sQTL) was also performed for all 4 traits. N denotes the actual sample size of studies included in each analysis. Colocalization analysis was based on nominally significant proteins and transcripts for Smk and proteins for AUD. All significant proteins (after Bonferroni correction) were used as input for drug-gene interaction analysis for all 4 traits. Only proteome-wide significant proteins for Smk and AUD were included for protein-protein interaction analysis.
