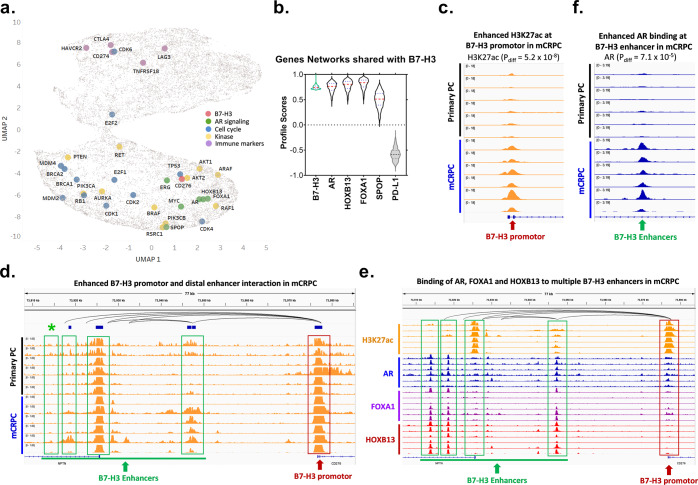
Fig. 2. B7-H3 is significantly associated with and regulated by AR-signaling pathways.
a Machine-learning (ML)-based UMAP analysis of the association between B7-H3 and key PC pathways. Each dot in UMAP represents one gene. The spatial distance between two genes represents the similarity of their gene networks. Key PC pathways are visualized including AR signaling (green), Cell cycle (blue), Kinases (yellow), and Immune markers (purple) along with B7-H3 (pink). b ML-based analysis of the gene-network association between B7-H3 and key AR-signaling pathway genes in mCRPC patients (SU2C/PCF, n = 208). Data are shown in violin plots, in which red lines represent median and blue lines represent first quartile (lower) and third quartile (upper). The boundary of the violin represents the range of all data points. The degree of overlap of the plots represents the similarity of the networks they are associated with. PD-L1 was used as a negative control. c Comparison of H3K27ac enhancement at B7-H3 promoter in mCRPC and primary PC from representative patient-derived xenografts (PDXs)22. Pdiff indicates the FDR-adjusted P-value for comparison between primary prostate cancer and mCRPC using DESeq2. d Enhanced interaction between B7-H3 distal enhancers (green box) and its promoter (red box) in mCRPC as compared to primary PC. Top track indicates H3K27ac HiChIP data from LNCaP, which reflects long range chromosomal interactions in LNCaP cells. e Binding of AR, FOXA1, and HOXB13 to multiple putative B7-H3 enhancer sequences (green box) in mCRPC. f Increased AR binding in mCRPC at one of the putative CD276 enhancers indicated in d, labeled with green *. Six representative AR ChIP-seq profiles of primary prostate cancer and mCRPC are shown32.