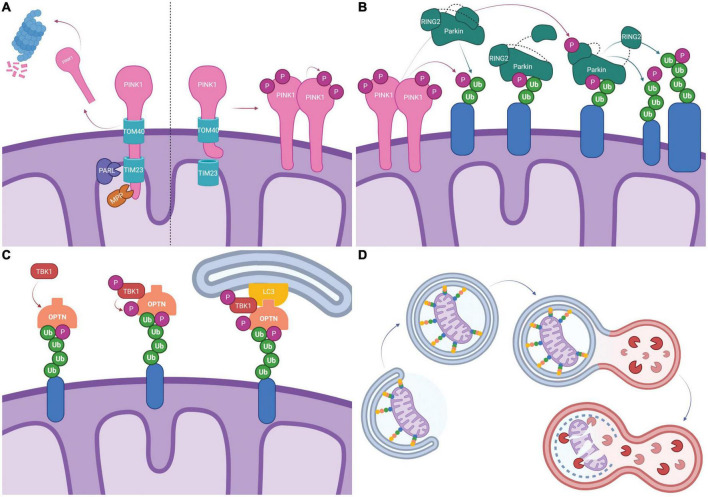
FIGURE 1.
Schematic representation of PINK1/Parkin dependent mitophagy. (A) Typically, PINK1 is imported into the mitochondria by TOM40/TIM23 where it is cleaved by mitochondrial proteases PARL and MPP. The c-terminal PINK1 fragment dissociates and is degraded by the proteasome (Left). Following mitochondrial depolarization, PINK1 import via TIM23 is blocked by electrostatic repulsion. PINK1 disassociates from TOM40, forms dimers and is activated by auto-phosphorylation (Right). (B) Activated PINK1 phosphorylates ubiquitinated mitochondrial proteins (shown in blue). Parkin binds phosphorylated ubiquitin which causes a conformational change and exposes the UBL allowing it to be phosphorylated by PINK1. This leads to additional conformational changes and exposes the catalytic site on RING2, fully activating Parkin. Parkin then ubiquitinates mitochondrial proteins, creating more substrates for PINK1 phosphorylation and feed-forward signal amplification. (C) Mitophagy receptors, here represented by OPTN, bind ubiquitinated mitochondrial proteins (shown in blue). TBK1 mediated phosphorylation of mitophagy receptors enhances ubiquitin binding. LIR domains on mitophagy receptors recruit the phagophore via LC3 binding. (D) Elongation of the phagophore leads to the engulfment of mitochondria into mitophagosomes. Mitophagosomes fuse with lysosomes and lysosomal proteases degrade the mitochondria. Created with BioRender.com.