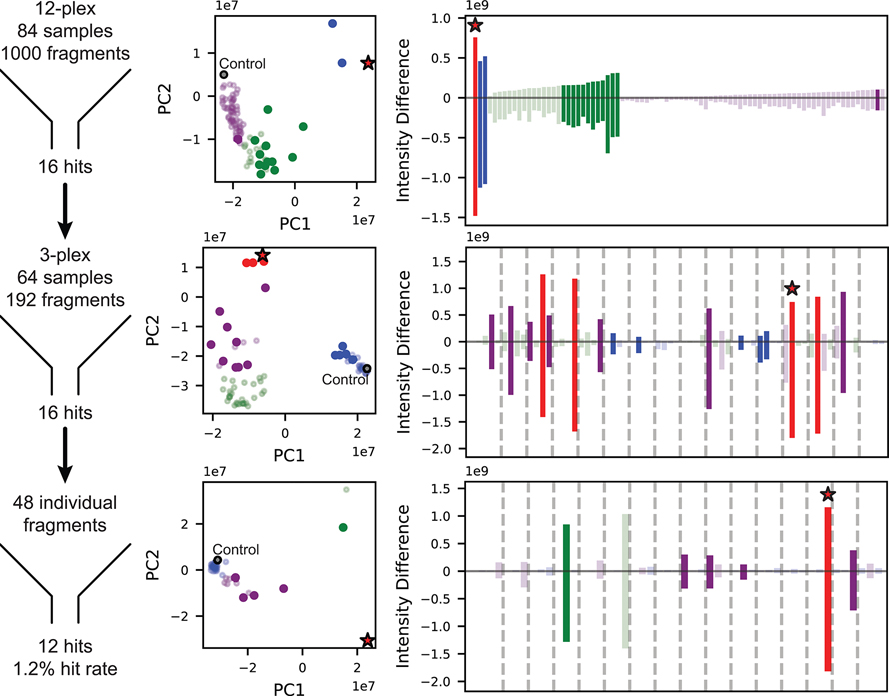
Figure 2.
Summary of protein-detected NMR-based fragment screen of Maybridge Library targeting PBRM1-BD2. The 1000 fragments were screened in pools of 12, 3, or individual compounds, and hits were identified through stepwise parsing of selected samples, yielding a final hit rate of 1.2% (left column). At each stage of parsing, identification of hit samples was aided by principal component analysis (middle column) and difference intensity analysis (right column) of the 2D HMQC spectra. Samples are colored according to k-means clustering of principal components. Throughout the screening process, samples selected as hits are represented as solid circles and bars, control samples in the PCA plots as gray circles outlined in black, and samples containing 5 are marked with a red star.