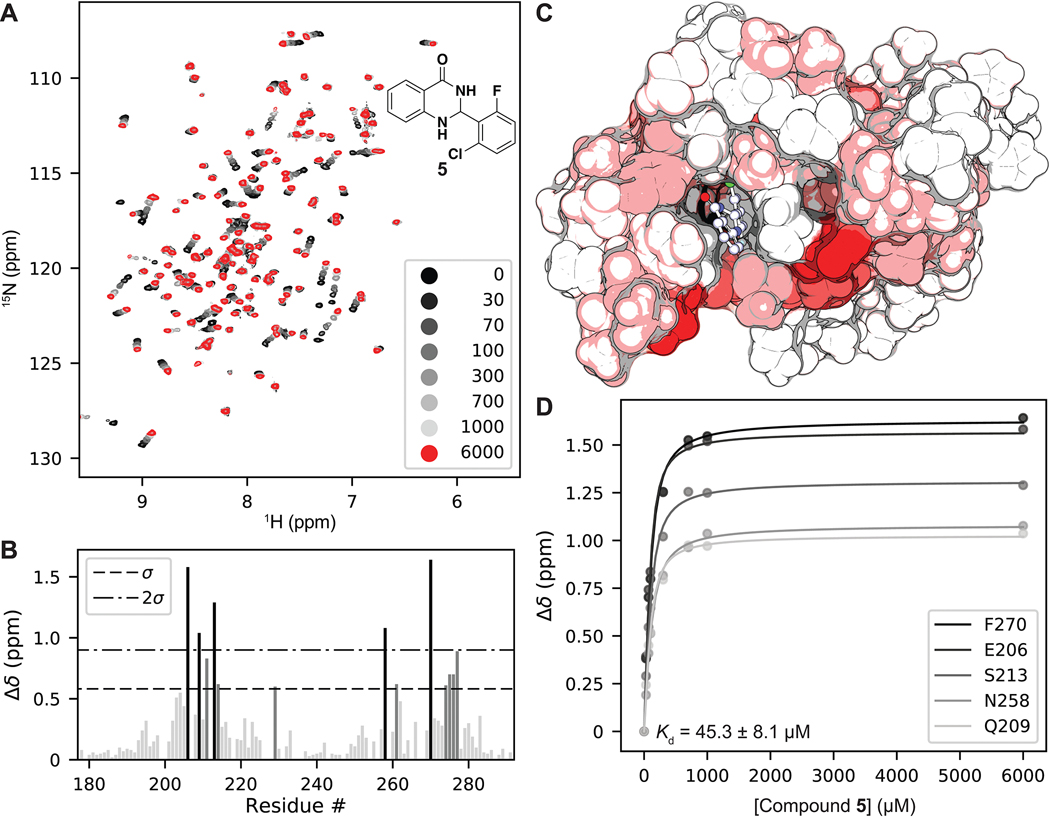
Figure 3.
(A) 1H, 15N SOFAST-HMQC overlays of PBRM1-BD2 titrated with increasing concentrations of 5 (structure in insert). (B) Quantification of total chemical shift perturbations (CSPs) (1H/15N Δδ chemical shift) manifested by 5 (6 mM) for individual amino acid residues of PBRM1-BD2. (C) Mapping of substantially perturbed residues on the crystal structure of PBRM1-BD2 (PDB 3LJW). The in silico docked pose of 5 into the acetyl-lysine binding site of PBRM1-BD2 is included. Residues displaying CSPs > 2σ (red), between 1σ and 2σ (pink), or <1σ (white) are indicated. (D) Concentration–response curves of indicated residues used to calculate binding affinity (Kd = 45.3 ± 8.1 μM) of 5 for PBRM1-BD2 using GraphPad Prism.