Abstract
Spatial patterning of neural stem cell populations is a powerful mechanism by which to generate neuronal diversity. In the developing Drosophila medulla, the symmetrically dividing neuroepithelial cells of the outer proliferation center crescent are spatially patterned by the nonoverlapping expression of 3 transcription factors: Vsx1 in the center, Optix in the adjacent arms, and Rx in the tips. These spatial genes compartmentalize the outer proliferation center and, together with the temporal patterning of neuroblasts, act to diversify medulla neuronal fates. The observation that the dorsal and ventral halves of the outer proliferation center also grow as distinct compartments, together with the fact that a subset of neuronal types is generated from only one half of the crescent, suggests that additional transcription factors spatially pattern the outer proliferation center along the dorsal-ventral axis. Here, we identify the spalt (salm and salr) and disco (disco and disco-r) genes as the dorsal-ventral patterning transcription factors of the outer proliferation center. Spalt and Disco are differentially expressed in the dorsal and ventral outer proliferation center from the embryo through to the third instar larva, where they cross-repress each other to form a sharp dorsal-ventral boundary. We show that hedgehog is necessary for Disco expression in the embryonic optic placode and that disco is subsequently required for the development of the ventral outer proliferation center and its neuronal progeny. We further demonstrate that this dorsal-ventral patterning axis acts independently of Vsx1-Optix-Rx and thus propose that Spalt and Disco represent a third outer proliferation center patterning axis that may act to further diversify medulla fates.
Keywords: Drosophila, optic lobe, Spalt, Disco, neuroepithelium, spatial patterning, compartment, medulla, neural stem cells
Introduction
The development of a complex nervous system requires that stem cells generate a diverse array of neurons in the correct order, location, and number. As a first step, neural stem cells are spatially patterned by signaling molecules and transcription factors, which assign them (and their progeny) unique positional identities (Holguera and Desplan 2018; Contreras et al. 2019; Chen and Konstantinides 2022). In the Drosophila nervous system, the spatial patterning of neural stem cells (termed neuroblasts) has been best studied in the embryonic ventral nerve cord, where the combinatorial action of segment polarity, columnar and Hox genes pattern neuroblasts along the anterior-posterior (A-P) and dorsal-ventral (D-V) axes (Doe 1992; Hirth et al. 1998; Bhat 1999; Lin and Lee 2012). Similarly, in the developing vertebrate neural tube, localized morphogen activity establishes distinct compartments of transcription factor expression, which confer unique identities to neural progenitors along both the A-P and D-V axes (Briscoe et al. 2000; Dasen et al. 2003; Sagner and Briscoe 2019). In both systems, spatially patterned progenitors are also temporally patterned by the sequential expression of transcription factors as they age (Kambadur et al. 1998; Isshiki et al. 2001; Kohwi and Doe 2013; Doe 2017; Delile et al. 2019). These 2 axes, spatial and temporal, work together to regulate multiple aspects of neurogenesis, including the generation of neural diversity (Delile et al. 2019; Sagner and Briscoe 2019; Sen et al. 2019; Rossi et al. 2021).
In recent years, the Drosophila optic lobe, comprised of the lamina, medulla, and lobula complex, has emerged as a powerful model system in which to study how the patterning of stem cells contributes to the development of a complex retinotopic circuit (Fischbach and Dittrich 1989; Nériec and Desplan 2016; Contreras et al. 2019; Malin and Desplan 2021). The largest structure in the optic lobe is the medulla, which mediates both motion and color processing and is comprised of >120 neuronal types (Fischbach and Dittrich 1989; Nériec and Desplan 2016; Özel et al. 2021). The medulla develops from a crescent of neuroepithelial (NE) cells termed the outer proliferation center (OPC; Egger et al. 2007; Li et al. 2013; Nériec and Desplan 2016; Erclik et al. 2017). In the third instar larva, the NE cells of the OPC are converted into neuroblasts, which then asymmetrically divide to generate the neurons and glia of the medulla (Ceron et al. 2001; Egger et al. 2007; Yasugi et al. 2008; Egger et al. 2010). Previous studies have shown that both temporal and spatial patterning inputs are required for medulla neuron specification (Hasegawa et al. 2011; Li et al. 2013; Erclik et al. 2017). In the temporal axis, the sequential expression of transcription factors in aging neuroblasts defines up to 11 distinct windows of specification (Hasegawa et al. 2011; Li et al. 2013; Konstantinides et al. 2022; Zhu et al. 2022). In the spatial axis, the OPC NE is patterned by the nonoverlapping expression of 3 homeobox transcription factors; Vsx1 in the center (cOPC), Optix in the adjacent arms (mOPC), and Rx in the tips (pOPC) (Fig. 1a; Gold and Brand 2014; Erclik et al. 2017). The Rx compartment is additionally subdivided by the restricted expression of the Dpp and Wg signaling genes (Kaphingst and Kunes 1994; Erclik et al. 2017). Neuroblasts integrate both temporal and spatial inputs to generate distinct neuronal types. For example, Pm3 neurons are specifically generated from neuroblasts derived from the Vsx1 spatial compartment and Hth temporal window (Erclik et al. 2017).
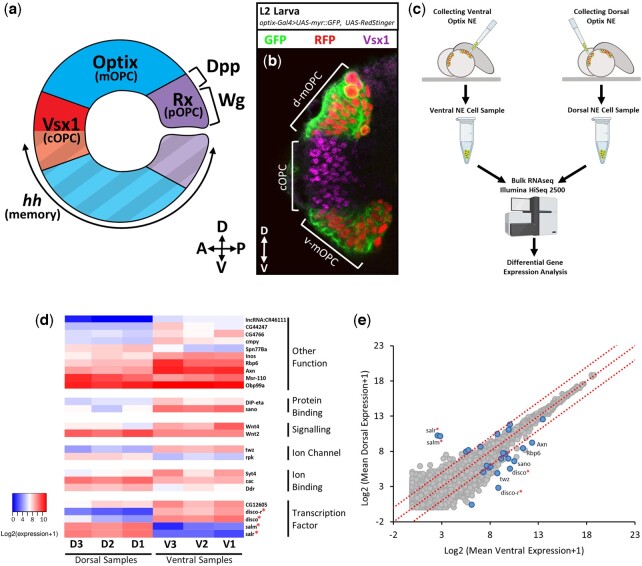
Fig. 1.
Identification of differentially expressed genes along the D-V axis of the OPC. a) A schematic of the spatial compartmentalization of the larval OPC NE. A memory trace of embryonic hh expression labels the ventral OPC. b) optix-Gal4 drives the expression of GFP (green) and RFP (red) in the dorsal and ventral mOPC compartments. c) Diagram of the workflow used to collect and sequence dorsal and ventral OPC NE cells. d) Log-transformed expression level heatmap of the 24 genes expressed at significantly different levels in the 3 dorsal (D1-3) vs 3 ventral (V1-3) cell samples (DESeq2, Wald test, adjusted P-value<0.05). Genes are organized and annotated according to their GO annotation protein functions. Red asterisks indicate the spalt and disco genes. e) Gene expression correlation plot of all genes expressed in the dorsal vs ventral NE cell samples. The mean expression level for each gene across the 3 replicates was log2 transformed and plotted. Points in blue are genes identified to have significantly different expression levels in the dorsal vs ventral samples (Wald test, adjusted P-value <0.05). Points labeled with gene names were upregulated in the dorsal or ventral samples with a log2 fold change >1.5.
In addition to the Vsx1-Optix-Rx spatial patterning of the OPC, several pieces of evidence suggest that a second spatial patterning mechanism compartmentalizes the OPC NE into dorsal and ventral halves. First, neurons with identical spatial and temporal addresses can assume different fates based on whether they are born in the dorsal or ventral half of the OPC; Pm1 and Pm2 neurons are each born from the Hth temporal window and Rx spatial compartment, but Pm1 neurons (marked by the expression of the Teashirt transcription factor) are born ventrally, whereas Pm2 neurons are generated dorsally (Erclik et al. 2017). Several other dorsal- and ventral-specific cell types have since been identified, including the ventrally derived neurons pDm8, yDm8, Tm4v, and Tm9v, and the dorsally derived neurons DRADm8, Tm4d, and Tm9d (Courgeon and Desplan 2019; Özel et al. 2021). Second, memory-trace experiments demonstrate that the dorsal and ventral halves of the OPC grow as distinct compartments; the NE cells labeled by the memory of hedgehog (hh) expression are strictly ventral and do not mix with unlabeled, dorsal cells (Erclik et al. 2017). Consequently, a sharp boundary is formed in the central Vsx1+ compartment between the ventral hh-lineage-marked cells and the dorsal unlabeled cells (Erclik et al. 2017). Of note, hh expression in the developing OPC is restricted to a short window in the early embryonic optic anlage, days before neurogenesis commences (Chang et al. 2001; Biehs et al. 2010). Thus, hh is unlikely to directly control neuronal fate specification. To date, transcription factors that are differentially expressed in the dorsal and ventral halves of OPC NE during larval development have not been reported.
Here, we identify the spalt (salm and salr) and disconnected (disco and disco-r) genes as dorsal- and ventral-specific OPC transcription factors, respectively. Spalt and Disco are differentially expressed in the dorsal and ventral OPC from the embryo through to the third instar larva, where they cross-repress each other to form a sharp D-V boundary. We show that disco is necessary for ventral fates, as it is required for both the development of the ventral OPC NE and the generation of ventrally derived neurons. We additionally show that embryonic hh is required for Disco expression in the optic placode. Taken together, our findings demonstrate that the Spalt–Disco patterning of the OPC represents a third patterning axis (independent of Vsx1-Optix-Rx spatial patterning) that may act to further diversify neuronal fates in the medulla.
Materials and methods
Micropipette cell isolation
To collect dorsal and ventral Optix+ OPC cell samples for bulk-RNA sequencing, we adapted a micropipette-based cell isolation protocol previously used to collect NE cells and neuroblasts (Caygill et al. 2012). To prepare the micropipettes, Borosilicate glass capillaries were pulled and beveled to achieve an inner needle diameter of 12 μm and tip angle of 45°. The inside of each micropipette was also coated with a siliconizing reagent (Sigmacote, Sigma-Aldrich) to prevent the collected cells from sticking to the inside of the micropipette. The coated needles were rinsed with water and left to dry overnight before use.
Female late second instar larvae of the genotype, UAS-RedStinger;Optix-Gal4;UAS-myr::GFP, were collected and the brains were dissected in 1xPBS. Brains were transferred to a Petri dish filled with 1xPBS and Kwik-Sil silicone adhesive was used to adhere the brains to the bottom of the dish, in an anterior mount orientation (Arain et al. 2021). The brains were visualized under a fluorescent scope fitted with a micromanipulator, onto which the preprepared microcapillary needle was mounted. Using the micromanipulator, the needle was brought into contact with the fluorescently labeled cells of one of the Optix+ OPC compartments. Using a syringe connected to the micropipette via a tubing system, the cells were drawn up into the pipette and expelled into a PCR tube prefilled with ∼5 μL of PBS. The tube was immediately flash frozen on dry ice to inhibit RNase activity. Cell sample collection occurred for a maximum period of 1 h after dissection, during which cells from either the dorsal or ventral compartment were collected from 3 to 4 brains and pooled into a PCR tube. Three dorsal and 3 ventral cell sample replicates were obtained for downstream sequencing analysis.
Bulk RNA-seq library preparation, sequencing, and transcriptome analysis
mRNA was extracted and purified using the SMART-Seq v4 Ultra Low Input RNA kit (Takara), and RNA-seq libraries were prepared using the Nextera XT Library Preparation Kit (Illumina). Sequencing was performed on the HiSeq 2500 (Illumina) system. The above library preparation and sequencing steps were performed by SickKids: The Centre for Applied Genomics.
A total of 40 million, 126 bp, paired-end, nondirectional reads were obtained per sample. The FASTQ data were first processed with Trimmomatic (version 0.36.6) for adaptor and low-quality base trimming (Bolger et al. 2014). RNA Star (version 2.6.0b-1) was used to align reads to the dm6 genome using the BDGP 6.31 genome assembly where ∼85% of reads were uniquely mapped (Dobin et al. 2013). From here, featureCounts (version 1.6.3+galaxy2) assigned ∼82% of the mapped reads to genes and the remainder were removed from downstream analysis due to low-confidence mapping (Liao et al. 2014). The read counts obtained by featureCounts were normalized between replicates and compared across each condition using DESeq2 (version 2.11.40.2) to identify upregulated genes in the dorsal and ventral samples (Love et al. 2014). All transcriptomic analyses described above were performed on the usegalaxy.org web platform (Afgan et al. 2018).
Antibody generation
A polyclonal antibody against Disco was generated by GenScript. The antibody was generated in guinea pigs using the following epitope:
>PYVMFGGQAGLHGLGLLSTGCQDPDSGSVDNEQDADPEDDNDFVYVDMQANSSSPAASSEDQEEHERDNEQDEEMHCSLSLASSSSIAADEERAADQPLDFSLHKRRKSEQDREQEQEQEQEREREAEKEQEQDVESDKEHEPEQEHELEREKRSPSDAFSMDQLLGKRKRHDSTASSSACSTAAASSASSSSASASANPPQTSIKMDLDPDSDSAYMTSRRQMLPLPVLDLEEHHHLRLLQTQMFAAAAAAAATSQAPPTAFLPAGSPVDLAKDSPPMWSLLSEMYRSMLLKTQ
Immunohistochemistry
Dissection and immunohistochemistry analysis of larval and adult brains were carried out using a previously published protocol (Arain et al. 2021). Briefly, brains were dissected in cold 1xPBS and fixed in 4% formaldehyde (in 1xPBS) for a total of 20 min at room temperature (for adult brains) or 30 min on ice (for larval brains). Brains were then washed 5 × in 400 μL of PBT (0.3% Triton in 1xPBS) before incubating in primary antibody solution overnight. After removal of the primary antibody solution, the brains were washed 5× in 400 μL of PBT and then placed on an orbital shaker to continue washing at room temperature for an additional 4 h. After the primary wash, the secondary antibody solution was added, and incubation occurred for 2 h at room temperature on an orbital shaker. Following the 2 h incubation, the antibody solution was removed and the brains were washed 5× with 400 μL PBT before being left immersed in PBT in a fridge overnight. Finally, brains were mounted in SlowFade (Invitrogen) mounting media before imaging.
Immunohistochemistry analysis of embryos was carried out using a previously published protocol with slight deviations (Kaczynski and Gunawardena 2010). Flies were placed in cages and left to lay on grape juice agar plates at 25°. Embryos were collected from the plates and rinsed in a fine sieve with deionized water. Embryos were then placed in a vial and immersed in 50% bleach solution (in water) for 3 min to dechorionate the embryos. Immediately after dechorionation, the embryos were washed well with deionized water. Following the wash, an equal parts heptane-fixative solution (4% formaldehyde in 1xPBS) was added before placing the vial on a nutator for 30 min to fix the embryos. Following fixation, the fixative layer of the heptane-fixative solution was removed and replaced with methanol. The vial was then shaken vigorously to remove the vitelline membranes of the embryos. Next, the heptane-methanol mixture was removed and embryos were rinsed in methanol for 5 × 5 min. Embryos were then rehydrated by washing them for 2 × 5 and 1 × 30 min in a 50% methanol, 50% PBT solution, then 1 × 30 min in 100% PBT on a nutator. Following rehydration, embryos were incubated with primary antibody solution in the fridge overnight. Embryos were then washed 10 × 6 min with PBT on a nutator. The secondary antibody solution was added and left to incubate for 2 h at room temperature on a nutator before washing 10 × 6 min with PBT on a nutator. Following the secondary antibody wash, embryos were submerged in Vectashield (Vector Labs) mounting media and left in the fridge overnight before mounting and imaging.
The following concentrations of primary antibodies were used to prepare the primary antibody solutions in PBT for a total volume of 100 μL (larvae and adult) or 50 μL (embryos): mouse anti-RFP (1:1,000; MBL International), chicken anti-GFP (1:1,000; Ivitrogen), rat anti-DE-Cadherin (1:20; DSHB), rabbit anti-Salm (1:100; gift from Claude Desplan), guinea pig anti-Disco (1:100; this study), mouse anti-Dac (1:10; DSHB), rat anti-Dpn (1:50; Abcam), rat anti-DN-Cadherin (1:20; DSHB), guinea pig anti-Vsx1 (1:500; Erclik et al. 2008), mouse anti-FasII (1:20; DSHB), goat anti-β-galactosidase (1:500; MP Biomedicals), mouse anti-Svp (1:200; DSHB), rabbit anti-Tsh (1:2,000; gift from Hector Herranz), rabbit anti-Lim1 (1:100; gift from Claude Desplan).
Secondary antibodies were prepared at 1:500 in PBT for a total volume of 100 μL (larvae and adult) or 50 μL (embryos). Secondary antibodies were obtained from Invitrogen and Jackson ImmunoResearch Laboratories.
Imaging and image analysis
Brains and embryos were imaged on a ZEISS LSM880 confocal microscope using a 25× oil objective. Images were processed using Volocity imaging software or ImageJ.
Fly strains and genetic crosses
Flies were reared at 25° on standard cornmeal food unless otherwise specified. OreR was used as the wild-type strain. The following fly stocks were used in this study: w*,UAS-myr::GFP;tubP-Gal80,FRT40A;tubP-Gal4/Tm6B (BDSC #5192, modified for this study), w*,hsFlp,tubP-Gal80,FRT19A;UAS-CD8::GFP;tubP-Gal4/Tm6B (BDSC #5134, modified for this study), hsFlp;;Act-FRT-Stop-FRT-Gal4,UAS-RFP (gift from Dorothea Godt), y1, w*,hsFlp;Df(2L)32FP5,FRT40A/cyo;MKRS/Tm6B (BDRC #29717), w*,Df(1)ED7355,FRT19A/Fm7a (BDSC #8899, modified for this study), UAS-salm.K (BDSC #29715), UAS-disco.M (BDSC #78348), UAS-disco-r.M (BDSC #78342), w*;UAS-discoRNAi (VDRC #330175), UAS-Vsx1RNAi (BDSC #50684), y1, v1;;UAS-OptixRNAi (BDSC #31910), hh-Gal4,y1w*;UAS-Flp.D;Act-FRT-Stop-FRT-lacZ,w*;Optix-Gal4, w*; Gal80ts; pbx-Gal4, UAS-myr::GFP (gift from Claude Desplan), disco-rC50.1S1 (disco-r-LacZ, BDSC #78347),;; ry506, hhAC/Tm3 (BDSC #1749),;; ry506, hhP30 (hh-lacZ, BDSC #5530), disco1 (BDSC #5682), Bs, Dp(1; Y)BSC228 (BDSC #32518), and w*,MZVUM-Gal4;UAS-CD8::GFP (Erclik et al. 2008).
Genetic knock-out clones of the salm and disco gene pairs were generated using the MARCM system (Luo and Wu 2006). To generate salm gene deficiency clones, females of the genotype, w*,UAS-myr::GFP;tubP-Gal80,FRT40A;tubP-Gal4/Tm6B, were crossed to, y1,w*,hsFlp;Df(2L)32FP5,FRT40A/cyo;MKRS/Tm6B males. Progeny was heat-shocked at the second instar larval stage for 1 h at 37°C and dissected in the late third instar stage. To generate disco gene deficiency clones, females of the genotype, w*,Df(1)ED7355,FRT19A/Fm7a were crossed to w*,hsFlp,tubP-Gal80,FRT19A;UAS-CD8::GFP;tubP-Gal4/Tm6B males. Progeny of this cross were heat-shocked at the second instar larval stage for 2 h at 37°C and left to recover at room temperature for 1 h before applying a second heat shock for an additional 2 h. All larvae were dissected in the early or late third instar stage.
Salm, Disco, and Disco-r overexpression clones were made by crossing, hsFlp;;Act-FRT-Stop-FRT-Gal4,UAS-RFP virgin females to UAS-salm.K, UAS-disco.M, or UAS-disco-r.M males. Progeny were heat-shocked at the early second larval instar stage for 12 min at 37°C and left to recover for 48 h before dissection.
vsx1, optix, and disco RNAi knock-down clones were made by crossing hsFlp;;Act-FRT-Stop-FRT-Gal4,UAS-RFP virgin females to UAS-Vsx1RNAi, y1,v1;;UAS-OptixRNAi or w*;UAS-disco-rRNAi males. Progeny were heat-shocked at the early second larval instar stage for 12 min at 37°C and left to recover for 48 h before dissection.
Analysis of optic lobe scRNA-seq dataset
salm and disco expressing cell types in the adult optic lobe were identified using a published single-cell sequencing dataset on the adult optic lobe (GEO accession: GSE142789; Özel et al. 2021). The preclustered Seurat object (Adult.rds, GSE142787) was analyzed using the assigned cluster identities located in the “FinalIdents” field of the metadata. salm and disco expression in the clusters was calculated and differential gene expression analysis was performed using the FindAllMarkers function in Seurat 3.1.5 under default parameters (Wilcoxon rank-sum test) on nonintegrated gene expression.
Results
Identification of differentially expressed D-V patterning genes in the OPC
To identify genes that are differentially expressed along the D-V axis of the OPC, we transcriptionally profiled NE cells isolated from the dorsal and ventral Optix+ (mOPC) compartments. We chose to collect cells from the Optix+ compartments because the compartments are large, and, unlike the dorsal and ventral Vsx1+ compartments, exist as spatially distinct cell populations (Fig. 1, a and b and Supplementary Fig. 1a). To isolate NE cells, we modified a protocol that was previously developed to extract fluorescently labeled cells from live larval brains using a glass micropipette (Fig. 1c; Caygill et al. 2012). Fluorescent labeling of the dorsal and ventral compartments was achieved with optix-Gal4 driving the expression of a nuclear RFP and membrane-bound GFP in the mOPC (Fig. 1b). Fluorescent cells were extracted via micropipette and 3 replicates each of dorsal and ventral cells were sent for Illumina mRNA sequencing (Fig. 1c).
Differential gene expression analysis between the dorsal and ventral mOPC samples revealed that the cells from these 2 compartments have very similar transcription profiles. mOPC genes (optix, combgap) were highly enriched in all samples compared to cOPC (vsx1, pxb) and pOPC (rx, dpp) genes, confirming that the isolated cells were collected from the Optix+ compartments (Supplementary Fig. 1b). Comparative analysis of the cell sample gene expression profiles identified 24 genes that had significantly different expression levels between the dorsal and ventral samples (Wald test, P < 0.05; Fig. 1d). Eight of these genes displayed large expression level differences between the dorsal vs ventral cells, with a log2 fold-change greater than 1.5 (Fig. 1e). One ventral-specific gene that was identified was DWnt4, which encodes a secreted signaling protein. DWnt4 expression has been previously shown to be restricted to the ventral OPC lamina precursor cells (LPC; Sato et al. 2006). The differential expression of DWnt4 in our ventral samples thus serves as confirmation that our cell extraction technique reliably isolated ventral vs dorsal cells. Gene Ontology (GO) annotation analyses identified several classes of differentially expressed D-V genes, including signaling proteins and transcription factors (Fig. 1d). Strikingly, 4 of the genes displaying the greatest expression level differences between the dorsal and ventral samples were comprised of 2 pairs of related zinc finger transcription factors: (1) spalt major (salm) and spalt-related (salr) (together referred to as the spalt genes) expressed in dorsal samples, and (2) disconnected (disco) and disco-related (disco-r) (together referred to as the disco genes) expressed in ventral samples (Fig. 1, d and e). The complementary expression of these genes in the dorsal and ventral samples, together with their previously reported roles in patterning developing fly appendages (Cohen et al. 1991; Patel et al. 2007; Grieder et al. 2009; Organista and De Celis 2013), led us to investigate whether the genes confer D-V identities to the NE cells of the developing OPC.
Salm and Disco expression divides the OPC NE into dorsal and ventral domains
As a first step in understanding the roles of the spalt and disco genes in OPC development, we performed immunohistochemistry analysis to confirm that the transcription factors are differentially expressed along the D-V axis of the OPC. Immunostaining of early third instar larval brains with Salm and Disco antibodies was consistent with our transcriptomic data; Salm is specifically expressed in the dorsal OPC (d-OPC) NE and Disco is reciprocally expressed in the NE of the ventral OPC (v-OPC; Fig. 2, a and b). The D-V pattern of Salm/Disco expression extends to all OPC compartments (Vsx1, Optix, and Rx) and the expression of the 2 genes meets in the cOPC, where they form a sharp boundary (Fig. 2, a and b and Supplementary Fig. 2a). Of note, Salm is expressed at lower levels in the cOPC relative to the other dorsal OPC compartments (Fig. 2a), which suggests that the spalt genes may play a compartment-specific role in the Vsx1 region. In addition, the disco-rC50.1S1 lacZ enhancer trap line, which reports for disco-r expression (Patel et al. 2007), is expressed in the ventral compartment of the OPC NE, consistent with the transcriptomic data that both disco paralogs are expressed in the ventral OPC (Supplementary Fig. 2b). We could not confirm that Salr is coexpressed with Salm in the dorsal OPC, as neither an antibody nor reporter line specific to the gene is currently available. The restricted D-V expression pattern of Salm and Disco was also observed in earlier larval stages, but not in late third instar brains, where only Salm continues to be expressed in the dorsal OPC (Fig. 2, c and d). Notably, Salm and Disco expression in the OPC is absent from the NE cells lateral to the furrow, which contribute to the lamina, and is restricted to the NE cells that give rise to the neuroblasts and neurons of the medulla (Fig. 2, e and f).
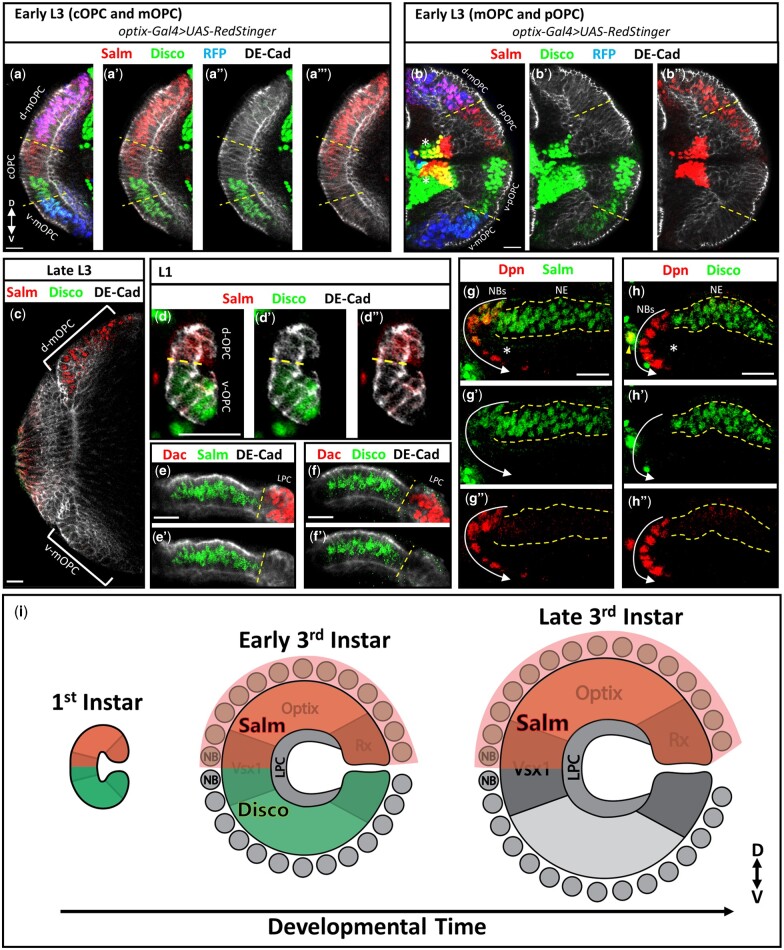
Fig. 2.
Salm and Disco are expressed in the dorsal and ventral halves of the OPC. a–b”) Salm (red) and Disco (green) expression in the OPC NE of the early third instar larva. NE is labeled by DE-Cadherin (DE-Cad, grey). Dashed lines indicate the mOPC-cOPC (a–a’’’) and mOPC-pOPC (b–b”) compartment boundaries as marked by optix-Gal4 driving the expression of RFP (blue). White asterisks (b-b”) mark Salm and Disco expression in pOPC-derived neurons. c–d”) Salm (red) and Disco (green) expression in the OPC NE (DE-Cad, grey) of late third instar (c) and first instar (d–d”) larvae. Yellow dashed lines (d-d”) indicate the location of the D-V expression boundary. e–f’) Expression of Salm (e–e’, green) and Disco (f–f’, green) in the OPC NE (DE-Cad, grey) of the early third instar larva is absent in the LPC as marked by Dachshund (Dac, red). Yellow dashed lines mark the lamina furrow separating the medial and lateral OPC NE. g–h”) Expression of Salm (g–g”, green) and Disco (h–h”, green) in neuroblasts (NBs) marked by Deadpan (Dpn, red) and neurons (asterisks) in the optic lobe of the early third instar larva. Arrows indicate the ages of the neuroblasts from the youngest to the oldest. Dashed lines outline the OPC NE. Yellow arrowhead (h) is a central brain neuroblast coexpressing Dpn and Disco. a–d”) Dorsal is up. e–h”) Medial is left. In all images: scale bar = 15μm. i) Cartoon schematic of Salm and Disco expression in the OPC NE, NBs, and LPC throughout larval development.
Next, we assessed whether Salm and Disco are expressed in other cells of the larval optic lobe. Within the developing medulla, Salm is additionally expressed in the youngest dorsal medulla neuroblasts, those closest to the NE, but is downregulated in older neuroblasts (Fig. 2g). In contrast, Disco is not expressed in medulla neuroblasts, indicating that it is downregulated during the NE to neuroblast transition (Fig. 2h). Neither gene is expressed in neurons derived from the OPC, apart from a subset of neurons born from the pOPC tips (Fig. 2, b, g, and h). In these pOPC-derived neurons, Salm and Disco do not exhibit D-V specificity, likely due to an additional independent role these factors play in the specification of Wingless-derived pOPC neurons (Bertet et al. 2014). Disco is also expressed in the NE cells of the ventral tip of the inner proliferation center, a progenitor population that gives rise to neurons of the lobula complex (Supplementary Fig. 2c; Hofbauer and Campos-Ortega 1990; Nériec and Desplan 2016).
Consistent with the absence of Salm and Disco expression in most OPC-derived neurons in the larva, these transcription factors are not expressed in the neurons of the adult medulla (Supplementary Fig. 2d). The genes are expressed in a subset of lobula complex neurons, which may represent the Salm and Disco expressing neurons derived from the larval pOPC (Supplementary Fig. 2d). To identify these neurons, we analyzed a previously published single-cell sequencing dataset of the whole adult optic lobe (Özel et al. 2021). This analysis identified 18 neuronal clusters with upregulated disco levels, including the neurons LC14, LLPC1, and LC16, and 5 clusters with upregulated salm levels, including LPLC1 and LPLC2 neurons (Wilcoxon Rank-Sum, P-adj < 0.05; Wu et al. 2016).
Taken together, the above results demonstrate that the expression of Salm and Disco subdivides the OPC NE into dorsal and ventral regions, respectively (Fig. 2i). The genes are expressed in all OPC compartments and form a sharp D-V boundary in the cOPC (Fig. 2i). We next determined whether these genes functionally interact to establish the D-V compartment boundary.
Spalt and Disco cross-repress each other in the developing OPC
The spatial compartmentalization of the OPC along the A-P axis requires cross-repressive interactions between the Vsx1, Optix, and Rx transcription factors (Erclik et al. 2017; Islam et al. 2021). We therefore asked whether Salm and Disco also cross-repress each other in the OPC NE to maintain the D-V compartment boundary. Ectopic expression of salm is sufficient to repress Disco in the v-OPC NE (Fig. 3a). Similarly, misexpression of disco or disco-r is sufficient to repress Salm in the d-OPC NE (Fig. 3b and Supplementary Fig. 3a). To test for the necessity of these genes in repressing one another we generated clones deficient for both genes in each paralogous gene pair, since, in other tissues in which the salm/salr and disco/disco-r genes are also coexpressed, the genes are functionally redundant (De Celis et al. 1996; Mahaffey et al. 2001; Mollereau et al. 2001). We thus made use of deficiency lines with chromosomal deletions that span the coding regions of the spalt genes [Df(2L)32FP5] or the disco genes [Df(1)ED7355] (Barrio et al. 1999; Patel et al. 2007) to generate loss-of-function clones in the larval OPC. In MARCM clones removing the spalt genes, Disco is not derepressed in the d-OPC NE (Fig. 3c and Supplementary Fig. 3b). Conversely, in MARCM clones removing the disco genes, Salm is derepressed in the v-OPC NE (Fig. 3d and Supplementary Fig. 3c). It should be noted that the levels of ectopic Salm expression in these clones are lower than those observed in the wild-type d-OPC NE, which suggests that additional genetic mechanisms may act to repress Salm expression in the v-OPC. To determine whether the disco genes act redundantly in the ventral NE to repress Salm expression, we generated RNAi clones in which only the expression of the disco gene is knocked down. We found that Salm is not derepressed in these disco RNAi clones (Supplementary Fig. 3d), which suggests that disco and disco-r act redundantly in the v-OPC NE. Taken together, the above data indicate that the disco genes are necessary and sufficient to repress Salm in the OPC, whereas the spalt genes are only sufficient to repress Disco (Fig. 3f).
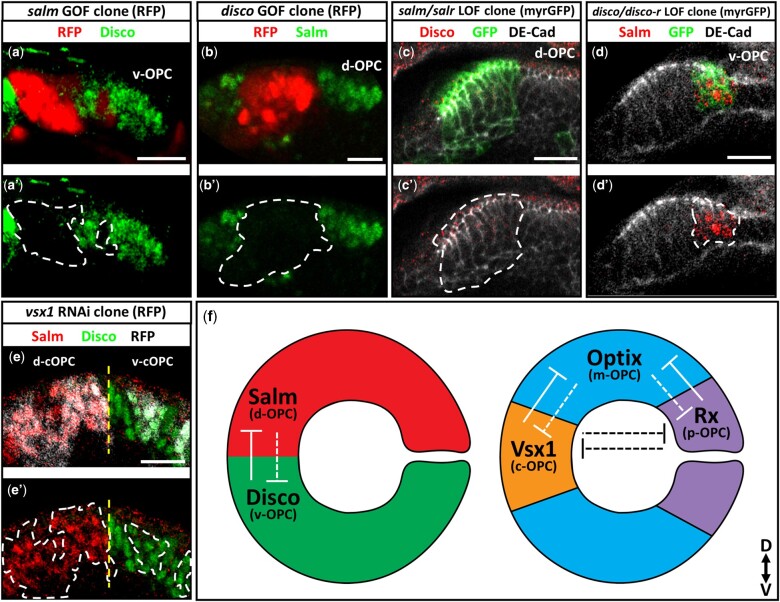
Fig. 3.
Salm and Disco cross-repress each other in the OPC. a–b’) salm (a–a’) and disco (b–b’) gain-of-function clones labeled by RFP (red) in the OPC NE. a–a’) Disco (green) expression in the v-OPC. (b-b’) Salm (green) expression in the d-OPC. c–d’) MARCM deficiency clones (GFP, green) for the spalt genes (c–c’) and disco genes (d–d’) in the OPC NE (DE-Cad, grey). c–c’) Disco (red) expression in the d-OPC. (d–d’) Salm (red) expression in the v-OPC. e–e’) Salm (red) and Disco (green) expression in vsx1 RNAi clones (RFP, grey) generated in the cOPC. Yellow dashed lines mark the D-V boundary within the cOPC. a’, b’, c’, d’, e’) Clones are outlined in white dashed lines. In all images: scale bar = 15μm. f) Schematic of the cross-repressive interactions between Salm and Disco. Cross-repressive relationships between other spatial patterning genes, Vsx1, Optix, and Rx, are also depicted as previously published (Erclik et al. 2017). Solid lines represent necessity and sufficiency. Dashed lines indicate sufficiency only.
We next determined whether the spalt and disco genes interact with the previously identified spatial patterning axis. Knockdown of vsx1 does not affect the expression of Salm or Disco in the cOPC and removal of the disco genes has no effect on Vsx1 expression (Fig. 3e and Supplementary Fig. 3e). Furthermore, knockdown of optix has no effect on Salm or Disco expression in the mOPC (Supplementary Fig. 3, f and g). These data suggest that the D-V patterning axis of the OPC is independent of the Vsx1-Optix-Rx spatial axis (Fig. 3f). To further investigate how the D-V axis is established, we next analyzed the regulatory relationships between spalt, disco, and vsx1 in the embryo.
hh is required for disco expression in the embryonic optic placode
The larval OPC is derived from a patch of ectodermal cells in the embryo termed the optic placode (Hartenstein and Campos-Ortega 1984; Daniel et al. 1999). Larval OPC patterning genes, such as vsx1 and wg, have been previously shown to establish their respective spatial compartments in the embryonic placode (Erclik et al. 2008; Hakes et al. 2018). Thus, we asked whether Salm and Disco expression also demarcate the dorsal and ventral halves of the future OPC in the optic placode. Costaining of embryos at various developmental stages with Salm, Disco, and the optic placode marker, Fasciclin II (Fas II), revealed that Salm and Disco subdivide the placode into 2 distinct domains as early as stage 11 and that this expression pattern is maintained throughout later embryonic stages (Fig. 4, a and b). Furthermore, as observed in the larval OPC, the expression of these genes forms a sharp boundary in the optic placode that is located within the Vsx1 domain, labeled by MZVUM-Gal4 (Fig. 4b; Erclik et al. 2008). We thus refer to the Salm and Disco expressing domains henceforth as the “dorsal” and “ventral” domains of the optic placode, respectively.
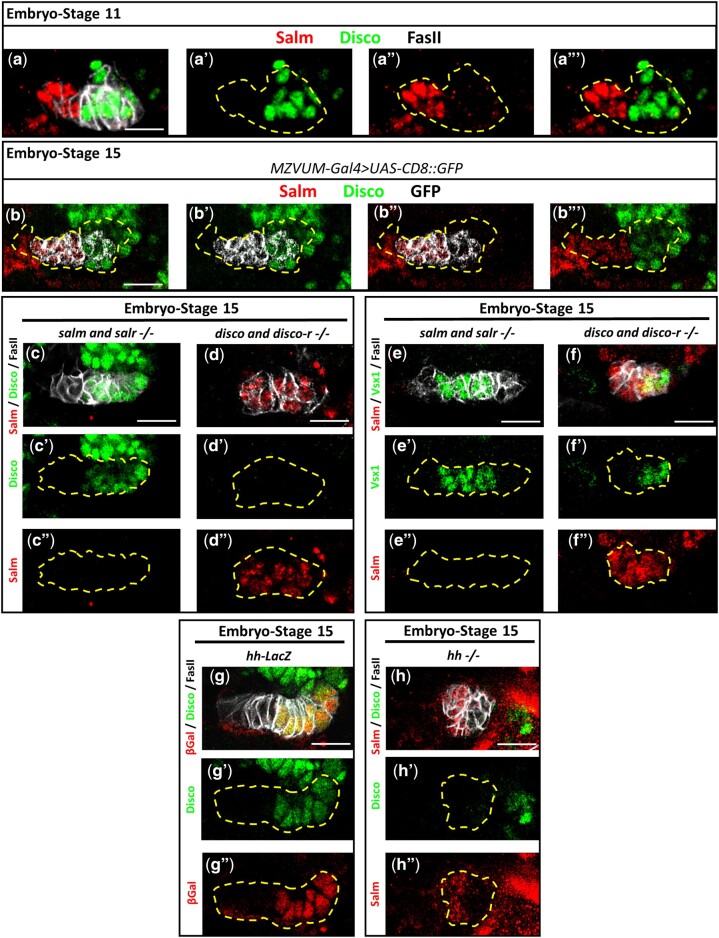
Fig. 4.
hh is required in the embryonic optic placode for Disco expression. a–b’’’) Salm (red) and Disco (green) are expressed in two distinct domains of the embryonic optic placode visualized at stage 11 (a–a’’’) and stage 15 (b–b’’’). b–b’’’) Salm and Disco establish a D-V boundary in the vsx1 expressing domain of the optic placode as labeled by MZVUM-Gal4 driving GFP (grey). c–f”) Salm (red) and Disco (c–d”, green) or Vsx1 (e–f”, green) expression in the stage 15 optic placode of whole mutant embryos deficient for the spalt genes (c–c”, e–e”) or disco genes (d–d”, f–f”). g–g”) Coexpression of Disco (green) and βGal (red) in the stage 15 optic placode of the hh lacZ enhancer trap line, hhP30. h–h”) Salm (red) and Disco (green) expression in the stage 15 optic placode of hh mutant embryos (hhAC). In all images: Cells of the optic placode labeled by FasII (grey in a, c, d, e, f, g, h, and not shown in b) are outlined with yellow dashed lines. Scale bar = 15μm.
We next determined whether the cross-repressive relationships observed between Salm and Disco in the larval OPC (Fig. 3i) are also present in the optic placode. We stained whole mutant embryos deficient for the spalt genes [Df(2L)32FP5] or disco genes [Df(1)ED7355] for Salm and Disco to determine the effects of losing these transcription factor pairs on optic placode patterning. As observed in the larval OPC, loss of the spalt genes does not affect Disco, which remains expressed in the ventral half of the placode in mutant embryos (Fig. 4c). Furthermore, Vsx1 expression in this genetic background is not affected (Fig. 4e). In contrast, in disco mutant embryos, all cells of the optic placode express Salm (Fig. 4d). We speculated that the Salm expression observed throughout the disco mutant placode could result from one of 2 mechanisms: (1) Derepression of Salm in the ventral half of the placode, or (2) Loss of the ventral placode, resulting in only Salm-positive dorsal cells remaining. To distinguish between these possibilities, we first determined where Vsx1 is expressed in disco mutants and found that Vsx1 is no longer expressed centrally, but rather at the margin of the placode (Fig. 4f). This observation supports our second proposed mechanism, as the localization of Vsx1 expression to the edge of the placode suggests that the ventral half of the placode is absent. Next, we compared the width and number of cells in disco mutant and wild-type placodes of stage 15 embryos and found that the disco mutant placode is significantly shorter (mean = 35.67 ± 7.4 μm in wild-type vs 27.24 ± 5.1 μm in disco, t-test, P < 0.05) and comprised of significantly fewer cells (mean = 24.78 ± 3 in wild-type vs 15.13 ± 3.1 in disco, t-test, P < 0.05) (Supplementary Figure 4 a and b). The observation that the disco mutant placode is significantly smaller than the wild-type placode further supports the model that the ventral placode is missing in disco mutants.
We next explored whether embryonic hh expression in the optic placode is required for Disco expression in the ventral half. Previous studies have shown that hh is expressed in a subset of optic placode cells as early as embryonic stage 11 before being downregulated in the late embryo (Chang et al. 2001; Biehs et al. 2010). The observation that a hh lineage trace labels the ventral half of the larval OPC (Erclik et al. 2017), led us to hypothesize that hh could be an upstream genetic regulator of Disco. As a first step, we made use of the enhancer trap line, hhP30, which contains a lacZ insertion in the hh locus, to report for hh expression (Lee et al. 1992). Costaining these embryos for βGal and Disco revealed that hh and Disco are indeed coexpressed in the optic placode (Fig. 4g). Furthermore, in the larva, the ventral OPC NE labeled by a hh memory trace coexpresses Disco, indicating that the Disco and hh expressing cells of the embryonic optic placode give rise to the ventral larval OPC (Supplementary Fig. 4c). We next tested whether hh is required for Disco expression using embryos homozygous for the amorphic allele, hhAC (Lee et al. 1992). In hh mutant embryos, Disco is downregulated in the ventral half of the optic placode (Fig. 4h). Interestingly, Salm expression in hh mutant embryos is unaffected and continues to be expressed in the dorsal half of the optic placode, despite the loss of Disco expression (Fig. 4h). Of note, we also observed that, in contrast to the elongated band of cells present in the wild-type placode, the cells within the hh mutant placode form a rosette-like compact structure (Fig. 4, b, g and h). Despite the altered structure, the number of cells in the optic placode of hh mutants is unaffected in comparison to the wild-type placode (mean = 24.78 ± 3 in wild-type vs 25.8 ± 2.3 in hhAC, t-test, P > 0.05; Supplementary Fig. 4b).
Taken together, the above data demonstrate that the D-V boundary of the OPC is established in the embryo, where hh is required for the ventral expression of Disco in the optic placode. In disco mutants, the ventral placode is absent and only dorsal Salm expressing cells remain. Given the severe disco loss-of-function phenotype in the optic placode, we next determined what role disco plays in the development of the larval OPC.
Disco is required for ventral OPC development
To analyze how the larval OPC develops from a disco mutant optic placode, we took advantage of a mutant allele, disco1, that fails to express Disco in the optic placode and OPC, but is normally expressed in other parts of the embryo (Heilig et al. 1991; Lee et al. 1999). In disco1 third instar larval brains, the OPC does not form the characteristic crescent shape observed in wild-type animals (Fig. 5, a–d). Remarkably, Salm is expressed throughout the disco1 mutant OPC NE and the Vsx1+ domain is located at the tip of the crescent (Fig. 5, b and c). These phenotypes are similar to those observed in the embryonic placode and suggest that the ventral OPC is missing in disco1 mutants. We confirmed that the ventral OPC phenotype maps to the disco locus using 2 genetic approaches: (1) disco1/Df(1)ED7355 heterozygous mutant larvae also fail to form the ventral OPC (Supplementary Fig. 5a), and (2) ventral OPC development is rescued in disco1 males carrying a Y-linked duplication that spans the disco genes (Supplementary Fig. 5b). Taken together, the embryonic and larval disco mutant phenotypes suggest that the disco genes are required for the development of the ventral placode/OPC. These results additionally confirm that Vsx1 does not require the disco genes for its expression.
Fig. 5.
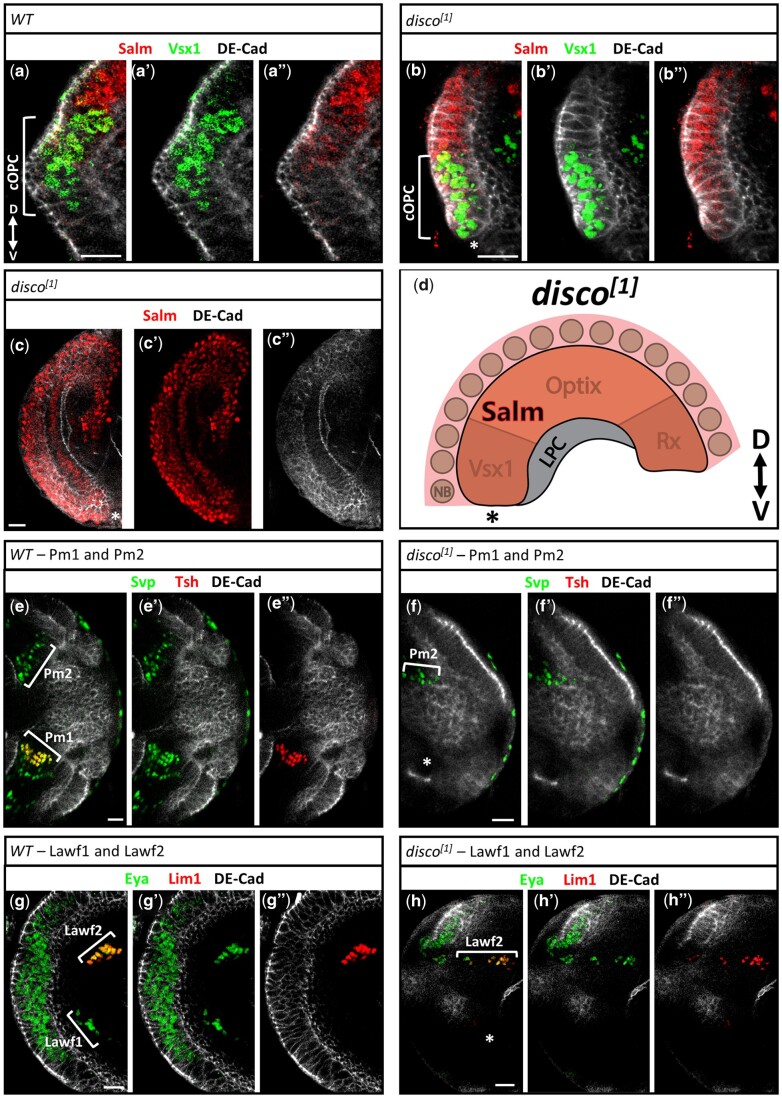
disco is required for the development of the ventral OPC and neurons. a–b”) Salm (red) and Vsx1 (green) in the cOPC NE of third instar wild-type (a–a”) and disco1 mutant (b–b”) larvae. A white asterisk (b) marks the ventral tip of the mutant OPC. c–c”) Salm (red) is expressed in the entirety of the OPC NE of a third instar disco1 mutant larva. A white asterisk marks the ventral tip of the mutant OPC. d) Cartoon schematic of the OPC NE of disco1 mutant larvae where the asterisk marks the ventral tip of the mutant OPC. e–f”) Pm1 and Pm2 neurons in the optic lobe of wild-type (e–e”) and disco1 mutant (f–f”) larvae labeled by Svp (green) and Tsh (red). A white asterisk (f) marks the expected location of Pm1 neurons. g–h”) Lawf1 and Lawf2 neurons in the optic lobe of wild-type (g–g”) and disco1 mutant (h–h”) larvae labeled by Eya (green) and Lim1 (red). A white asterisk (h) marks the expected location of Lawf1 neurons. In all images: Scale bar = 15μm. Dorsal is up. OPC NE is labeled by DE-Cad (grey).
We next determined whether medulla neurogenesis is affected in disco mutant brains. We analyzed disco1 larval brains for the generation of 2 sets of medulla neurons that are born on opposite sides of the D-V boundary. The first pair, Pm1 and Pm2, are generated from the Rx spatial region and Hth temporal window (Erclik et al. 2017). Pm1 neurons are born ventrally and express the TFs Seven up (Svp) and Teashirt (Tsh), whereas Pm2 neurons are born dorsally and only express Svp (Fig. 5e; Erclik et al. 2017). In the disco1 mutant medulla cortex, the ventral Pm1 population is absent, but the dorsal Pm2 cells are still specified (Fig. 5f). Analysis of a second pair of neurons gave a similar result. Lawf2 neurons, derived from the dorsal Rx region and marked by the TFs Eyes absent (Eya) and LIM homeobox 1 (Lim1; Chen et al. 2016; Suzuki et al. 2016), are still present in disco1 mutants, but ventral Lawf1 neurons, which express Eya only, are missing (Fig. 5, g and h). Although the absence of markers specific to Pm1 and Lawf1 is consistent with the possibility that these neurons are present but mis-specified in disco1 mutants, the finding that the ventral OPC does not develop in these mutants suggests that these neurons are never generated. Taken together, the above results demonstrate that disco is required for both the development of the ventral OPC and its neuronal output.
Discussion
In this study, we identify the spalt (salm and salr) and disco (disco and disco-r) paralogs as the D-V patterning transcription factors of the OPC. Salm (dorsal) and Disco (ventral) are differentially expressed in the OPC NE from the embryo through to the third instar larva, where the genes cross-repress each other to form a sharp D-V boundary in the cOPC. We show that this D-V patterning axis acts independently of the previously identified Vsx1-Optix-Rx spatial patterning axis. We also demonstrate that hh is necessary for disco expression in the embryonic optic placode and that disco is required for the development of the ventral OPC NE and its neuronal progeny. Taken together, our findings demonstrate that Spalt-Disco patterning of the OPC represents a third patterning axis (in addition to the previously identified temporal and spatial axes) that may act to diversify neuronal fates in the medulla.
Spalt and Disco: a second spatial patterning axis in the OPC
Previous hh lineage-trace experiments have demonstrated that the dorsal and ventral halves of the OPC grow as distinct compartments, with a sharp boundary in the Vsx1+ cOPC domain (Erclik et al. 2017). The observation that the expression of hh in the developing OPC is limited to a brief window in the early embryonic optic placode indicates that additional genes define the D-V compartments during larval development (Chang et al. 2001; Biehs et al. 2010). Our findings that Salm and Disco are specifically expressed in the growing dorsal and ventral compartments, respectively, and that they cross-repress each other to form compartment boundaries, suggest that these genes define the D-V patterning axis of the OPC.
Several pieces of evidence suggest that the Spalt-Disco patterning axis is established independent of the previously identified Vsx1-Optix-Rx spatial axis: (1) Neither Spalt nor Disco is required for the expression of Vsx1 in the embryonic optic placode, (2) Vsx1 is still expressed in the third instar larval OPC of disco1 mutants, and (3) neither Vsx1 nor Optix is required for the expression of Salm or Disco in the OPC NE. Future studies should extend this analysis to the Rx spatial compartment to further confirm that Spalt-Disco patterning is independent of the Vsx1-Optix-Rx patterning axis. Future analysis should also investigate whether 1 spatial axis is established before the other by determining when the Salm-Disco and Vsx1-Optix-Rx genes are first expressed in the embryonic optic placode. Vsx1 is expressed as early as stage 9 (Erclik et al. 2008) and we show here that both Salm and Disco are expressed at stage 11, but these genes may be expressed at earlier timepoints as well.
It will also be important to determine whether the spalt and disco genes act together with the existing spatial and temporal axes to diversify neuronal fates in the medulla. In recent years, several studies have identified neuronal cell types that are specifically generated in either the dorsal or ventral half of the medulla (Chen et al. 2016; Suzuki et al. 2016; Erclik et al. 2017; Courgeon and Desplan 2019; Özel et al. 2021). For example, Pm1 and Pm2 neurons are both born in the Hth temporal and Rx spatial windows. However, Pm1 neurons are generated ventrally, whereas Pm2 neurons are made dorsally (Erclik et al. 2017). We have shown here that Pm1 neurons are missing in disco mutant brains, but this phenotype is likely due to an embryonic role of disco in promoting the development of the ventral optic placode. Could spalt and disco also play a later role in the larval OPC NE to diversify the fates of neurons such as Pm1/2? And, if so, how are the 3 patterning axes (Vsx1-Optix-Rx spatial, D-V spatial, and temporal) integrated by neuroblasts and neurons to generate diversity? Patterning factors in other systems regulate specification by directly binding to enhancers to regulate transcription or by modifying chromatin accessibility landscapes to allow for the integration of additional inputs (Sen et al. 2019; Charest et al. 2020; Chen and Konstantinides 2022). The compartment-specific drivers available in the OPC, together with the relatively large number of spatially identical cells that can be isolated from larval brains, make the OPC an ideal system in which to use multiomic approaches to study how spatial and temporal inputs are integrated.
The D-V axis of the OPC is established in the embryo
Salm and Disco expression already subdivides the embryonic optic placode into the future ventral and dorsal halves of the OPC as early as stage 11, before the placode has invaginated from the ectoderm (Daniel et al. 1999). We show that hh is likely the factor that initiates Disco expression in the ventral half of the placode. The overlapping expression patterns of hh and Disco in the placode, together with the loss of Disco expression in hh mutant embryos, supports a model in which autocrine Hh signaling initiates Disco expression (Biehs et al. 2010). Hh autocrine signaling has been previously shown in the embryonic placode, where it activates the genes roadkill (rdx), snail (sna), and eya (Biehs et al. 2010). Surprisingly, we found that the ventral half of the optic placode is still present in hh mutant embryos, despite the loss of Disco expression. This phenotype is inconsistent with the disco LOF phenotype, in which the ventral placode is missing. We speculate that, although we did not detect Disco expression in hh mutant placodes, low levels of Disco may still be present, and that this residual expression is sufficient to promote ventral placode development. Alternatively, disco-r expression may be unaffected in the hh mutant placode, and thus, may function redundantly to direct ventral placode development.
Future studies should examine the embryonic placode at earlier stages of its specification to further understand how the D-V axis is established. For example, the observation that Salm is expressed in all cells of the early placode would support a model in which Hh signaling activates Disco to promote ventral fates. It would also be interesting to determine whether, in the early placode of disco mutant embryos, the ventral half is absent, which would indicate a failure in its specification, or if it is initially present and then subsequently lost at later embryonic stages.
Two distinct roles for disco in OPC development
We have found that the disco genes play 2 distinct roles in the developing OPC. Firstly, they are required in the embryo for the development of the ventral optic placode; in disco deficiency mutants, the placode is significantly smaller, Salm is expressed in all cells and Vsx1 expression is no longer centrally localized, but rather found at the margin. A similar phenotype is observed in the larval OPC of disco1 mutants, in which the ventral half of the crescent is absent, and we propose that both the embryonic and larval phenotypes are caused by a failure in ventral placode specification. The observation that the ventral OPC is missing in disco1 mutants, in which only the disco gene is mutated, is unexpected given that the disco genes act redundantly in other systems. We postulate that disco-r expression may also be lost in the embryonic placode of disco1 mutants. The point mutation in disco1 is located in one of Disco’s 2 zinc fingers and has been shown to specifically disrupt the ability of Disco to autoregulate its transcription in the optic placode (Lee et al. 1999). We speculate that this mutation may also affect the ability of Disco to bind to the enhancers required for the initiation of disco-r expression in the placode, which would result in a failure to express both genes.
The second role for the disco genes in the OPC is in the maintenance of D-V compartment boundaries, as the disco genes are both necessary and sufficient to repress Salm expression in the ventral OPC NE. As outlined above, we speculate that in addition to the maintenance of compartment identity, the disco (and spalt) genes may be required for the specification of the neuronal cell types derived from the NE. The proposed functions for spalt and disco are similar to the roles played by the Vsx1-Optix-Rx spatial patterning genes in the OPC NE and neurons. For example, Vsx1 is both required to maintain cOPC compartment identity by repressing Optix expression, and to specify the fates of neurons derived from the cOPC NE.
A conserved role for spalt and disco as D-V patterning genes
The spalt and disco gene pairs have previously been shown to be required for the specification of dorsal and ventral Drosophila appendages, respectively (Cohen et al. 1991; Patel et al. 2007; Grieder et al. 2009; Organista and De Celis 2013). In the wing disc (a dorsal appendage), spalt is required for various aspects of wing development, including the specification of the hinge tissue, and promotion of cell proliferation and survival (Grieder et al. 2009; Organista and De Celis 2013). Conversely, disco is expressed in the leg disc (a ventral appendage), where it is required for the development of distal leg segments (Cohen et al. 1991; Patel et al. 2007; Dey et al. 2009). Remarkably, misexpression of spalt in the leg disc or disco in the wing disc generates appendage transformations where the leg develops wing-like structures and the wing develops leg-like structures (Patel et al. 2007; Grieder et al. 2009). The developmental roles played by spalt and disco in D-V appendage specification may thus be evolutionarily linked to the function of these genes in assigning dorsal and ventral fates to the NE cells of the OPC.
The spalt and disco genes have also been implicated in other aspects of visual system development, though neither in a D-V specific manner. Spalt plays an essential role in multiple aspects of photoreceptor specification, including inner photoreceptor (R7 and R8) differentiation, while disco is required for the proper axonal guidance of the larval pioneer optic nerve, Bolwig’s organ (Steller et al. 1987; Mollereau et al. 2001). Of note, the adult optic lobe is disorganized in disco1 mutants, a phenotype that has been attributed to disco’s role in optic nerve targeting (Steller et al. 1987), but our data suggest that the optic lobe phenotype may also be due to the absence of the ventral OPC and its derivatives.
Intriguingly, sal-like protein 3 (SALL3), a vertebrate homolog of salm, has been demonstrated to play a role in the D-V specification of neurons in the developing mouse retina (Melo et al. 2011). Mutations in SALL3 disproportionately affect the specification of ventral horizontal cells, which suggests that there may be a conserved role for the spalt genes in the specification of visual system neurons along the D-V axis (Melo et al. 2011). It will thus be interesting to determine whether the disco vertebrate homolog, basonuclin (BNC) (Romano et al. 2004), also plays a role in vertebrate retinal development.
Supplementary Material
Acknowledgments
The authors would like to thank Claude Desplan, Dorothea Godt, and Hector Herranz for antibodies and fly stocks as well as Baohua Liu and Bryan Stewart for assistance with developing the cell isolation protocol. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and Vienna Drosophila Resource Center were used in this study. The DE-cad, DN-cad, Dac, FasII, and Svp monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242. Schematics were generated using Adobe Illustrator and BioRender.com.
Funding
This work was supported by an NSERC Discovery Grant (RGPIN2015-06457) awarded to TE. PV is supported by the Vision Science Research Program (University of Toronto: Ophthalmology and Vision Sciences and UHN), the Ontario Graduate Scholarship, and the Queen Elizabeth II/Pfizer Graduate Scholarship in Science and Technology.
Conflicts of interest
None declared.
Contributor Information
Priscilla Valentino, Department of Biology, University of Toronto Mississauga, Mississauga, ON L5L 1C6, Canada; Department of Cell and Systems Biology, University of Toronto, Toronto, ON M5S 1A1, Canada.
Ted Erclik, Department of Biology, University of Toronto Mississauga, Mississauga, ON L5L 1C6, Canada; Department of Cell and Systems Biology, University of Toronto, Toronto, ON M5S 1A1, Canada.
Data Availability
Drosophila strains and antibodies are available upon request. The raw and processed sequencing data used in this study have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE205934. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures.
Supplemental material is available at GENETICS online.
Literature cited
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arain U, Valentino P, Islam IM, Erclik T.. Dissection, immunohistochemistry and mounting of larval and adult Drosophila brains for optic lobe visualization. J Vis Exp. 2021;(170):e61273. doi: 10.3791/61273. [DOI] [PubMed] [Google Scholar]
- Barrio R, De Celis JF, Bolshakov S, Kafatos FC.. Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev Biol. 1999;215(1):33–47. doi: 10.1006/dbio.1999.9434. [DOI] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C.. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell. 2014;158(5):1173–1186. doi: 10.1016/J.CELL.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KM. Segment polarity genes in neuroblast formation and identity specification during Drosophila neurogenesis. Bioessays. 1999;21(6):472–485. doi:. [DOI] [PubMed] [Google Scholar]
- Biehs B, Kechris K, Liu SM, Kornberg TB.. Hedgehog targets in the Drosophila embryo and the mechanisms that generate tissue-specific outputs of Hedgehog signaling. Development. 2010;137(22):3887–3898. doi: 10.1242/dev.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J.. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101(4):435–445. doi: 10.1016/S0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Caygill EE, Gold KS, Brand AH.. Molecular profiling of neural stem cells in Drosophila melanogaster. In: Hassan BA, editor. The Making and Un-Making of Neuronal Circuits in Drosophila. Neuromethods. Vol. 69. Totowa (NJ: ): Humana Press; 2012. p. 249–260. [Google Scholar]
- Ceron J, González C, Tejedor FJ.. Patterns of cell division and expression of asymmetric cell fate determinants in postembryonic neuroblast lineages of Drosophila. Dev Biol. 2001;230(2):125–138. doi: 10.1006/DBIO.2000.0110. [DOI] [PubMed] [Google Scholar]
- Chang T, Mazotta J, Dumstrei K, Dumitrescu A, Hartenstein V.. Dpp and Hh signaling in the Drosophila embryonic eye field. Development. 2001;128(23):4691–4704. [DOI] [PubMed] [Google Scholar]
- Charest J, Daniele T, Wang J, Bykov A, Mandlbauer A, Asparuhova M, Röhsner J, Gutiérrez-Pérez P, Cochella L.. Combinatorial action of temporally segregated transcription factors. Dev Cell. 2020;55(4):483–499.e7. doi: 10.1016/J.DEVCEL.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Konstantinides N.. Integration of spatial and temporal patterning in the invertebrate and vertebrate nervous system. Front Neurosci. 2022;16:854422. doi: 10.3389/FNINS.2022.854422/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Del Valle Rodriguez A, Li X, Erclik T, Fernandes VM, Desplan C.. A unique class of neural progenitors in the Drosophila optic lobe generates both migrating neurons and glia. Cell Rep. 2016;15(4):774–786. doi: 10.1016/J.CELREP.2016.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B, Wimmer EA, Cohen SM.. Early development of leg and wing primordia in the Drosophila embryo. Mech Dev. 1991;33(3):229–240. doi: 10.1016/0925-4773(91)90030-A. [DOI] [PubMed] [Google Scholar]
- Contreras EG, Sierralta J, Oliva C.. Novel strategies for the generation of neuronal diversity: lessons from the fly visual system. Front Mol Neurosci. 2019;12:140. doi: 10.3389/fnmol.2019.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgeon M, Desplan C.. Coordination between stochastic and deterministic specification in the Drosophila visual system. Science. 2019;366(6463):eaay6727. doi: 10.1126/SCIENCE.AAY6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel A, Dumstrei K, Lengyel JA, Hartenstein V.. The control of cell fate in the embryonic visual system by atonal, tailless and EGFR signaling. Development. 1999;126(13):2945–2954. doi: 10.1242/DEV.126.13.2945. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu JP, Jessell TM.. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425(6961):926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- De Celis JF, Barrio R, Kafatos FC.. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996;381(6581):421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- Delile J, Rayon T, Melchionda M, Edwards A, Briscoe J, Sagner A.. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development. 2019;146(12):dev173807. doi: 10.1242/dev.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Zhao XL, Popo-Ola E, Campos AR.. Mutual regulation of the Drosophila disconnected (disco) and Distal-less (Dll) genes contributes to proximal-distal patterning of antenna and leg. Cell Tissue Res. 2009;338(2):227–240. doi: 10.1007/S00441-009-0865-Z/FIGURES/9. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116(4):855–863. doi: 10.1242/DEV.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Temporal patterning in the Drosophila CNS. Annu Rev Cell Dev Biol. 2017;12(55). doi: 10.1146/annurev-cellbio-111315. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/NAR/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ.. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH.. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137(18):2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erclik T, Hartenstein V, Lipshitz HD, McInnes RR.. Conserved role of the Vsx genes supports a monophyletic origin for bilaterian visual systems. Curr Biol. 2008;18(17):1278–1287. doi: 10.1016/j.cub.2008.07.076. [DOI] [PubMed] [Google Scholar]
- Erclik T, Li X, Courgeon M, Bertet C, Chen Z, Baumert R, Ng J, Koo C, Arain U, Behnia R, et al. Integration of temporal and spatial patterning generates neural diversity. Nature. 2017;541(7637):365–370. doi: 10.1038/nature20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach K-F, Dittrich APM.. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258(3):441–475. [Google Scholar]
- Gold KS, Brand AH. Optix defines a neuroepithelial compartment in the optic lobe of the Drosophila brain. Neural Dev. 2014;9(1):18–17. doi: 10.1186/1749-8104-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder NC, Morata G, Affolter M, Gehring WJ. Spalt major controls the development of the notum and of wing hinge primordia of the Drosophila melanogaster wing imaginal disc. Dev Biol. 2009;329(2):315–326. doi: 10.1016/j.ydbio.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Hakes AE, Otsuki L, Brand AH.. A newly discovered neural stem cell population is generated by the optic lobe neuroepithelium during embryogenesis in Drosophila melanogaster. Development. 2018;145(18):dev166207. doi: 10.1242/DEV.166207/VIDEO-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA.. Early neurogenesis in wild-type Drosophila melanogaster. Wilehm Roux Arch Dev Biol. 1984;193(5):308–325. doi: 10.1007/BF00848159. [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Kitada Y, Kaido M, Takayama R, Awasaki T, Tabata T, Sato M.. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development. 2011;138(5):983–993. doi: 10.1242/DEV.058370. [DOI] [PubMed] [Google Scholar]
- Heilig JS, Freeman M, Laverty T, Lee KJ, Campos AR, Rubin GM, Steller H.. Isolation and characterization of the disconnected gene of Drosophila melanogaster. EMBO J. 1991;10(4):809–815. doi: 10.1002/J.1460-2075.1991.TB08013.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F, Hartmann B, Reichert H.. Homeotic gene action in embryonic brain development of Drosophila. Development. 1998;125(9):1579–1589. doi: 10.1242/DEV.125.9.1579. [DOI] [PubMed] [Google Scholar]
- Hofbauer A, Campos-Ortega JA.. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Rouxs Arch Dev Biol. 1990;198(5):264–274. doi: 10.1007/BF00377393. [DOI] [PubMed] [Google Scholar]
- Holguera I, Desplan C.. Neuronal specification in space and time. Science. 2018;362(6411):176–180. doi: 10.1126/science.aas9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam IM, Ng J, Valentino P, Erclik T.. Identification of enhancers that drive the spatially restricted expression of Vsx1 and Rx in the outer proliferation center of the developing Drosophila optic lobe. Genome. 2021;64(2):109–117. doi: 10.1139/GEN-2020-0034. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106(4):511–521. doi: 10.1016/S0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Kaczynski TJ, Gunawardena S.. Visualization of the embryonic nervous system in whole-mount Drosophila embryos. J Vis Exp. 2010;(46):e2150. doi: 10.3791/2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF.. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12(2):246–260. doi: 10.1101/GAD.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaphingst K, Kunes S.. Pattern formation in the visual centers of the Drosophila brain: wingless acts via decapentaplegic to specify the dorsoventral axis. Cell. 1994;78(3):437–448. doi: 10.1016/0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ.. Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci. 2013;14(12):823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N, Holguera I, Rossi AM, Escobar A, Dudragne L, Chen YC, Tran TN, Martínez Jaimes AM, Özel MN, Simon F, et al. A complete temporal transcription factor series in the fly visual system. Nature. 2022;604(7905):316–322. doi: 10.1038/s41586-022-04564-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA.. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71(1):33–50. doi: 10.1016/0092-8674(92)90264-D. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Mukhopadhyay M, Pelka P, Campos AR, Steller H.. Autoregulation of the Drosophila disconnected gene in the developing visual system. Dev Biol. 1999;214(2):385–398. doi: 10.1006/dbio.1999.9420. [DOI] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C.. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013;498(7455):456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- Lin S, Lee T.. Generating neuronal diversity in the Drosophila central nervous system. Dev Dyn. 2012;241(1):57–68. doi: 10.1002/DVDY.22739. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Wu JS.. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006;1(6):2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- Mahaffey JW, Griswold CM, Cao Q-M.. The Drosophila genes disconnected and disco-related are redundant with respect to larval head development and accumulation of mRNAs from deformed target genes. Genetics. 2001;157(1):225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin J, Desplan C.. Neural specification, targeting, and circuit formation during visual system assembly. Proc Natl Acad Sci U S A. 2021;118(28). doi: 10.1073/pnas.2101823118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J, de Peng G-H, Chen S, Blackshaw S.. The Spalt family transcription factor Sall3 regulates the development of cone photoreceptors and retinal horizontal interneurons. Development. 2011;138(11):2325–2336. doi: 10.1242/DEV.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau B, Dominguez M, Webel R, Colley NJ, Keung B, De Celis JF, Desplan C.. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412(6850):911–913. doi: 10.1038/35091076. [DOI] [PubMed] [Google Scholar]
- Nériec N, Desplan C.. From the eye to the brain. development of the Drosophila visual system. Curr Top Dev Biol. 2016;116:247–271. doi: 10.1016/bs.ctdb.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organista MF, De Celis JF.. The Spalt transcription factors regulate cell proliferation, survival and epithelial integrity downstream of the Decapentaplegic signalling pathway. Biol Open. 2013;2(1):37–48. doi: 10.1242/BIO.20123038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özel MN, Simon F, Jafari S, Holguera I, Chen Y-C, Benhra N, El-Danaf RN, Kapuralin K, Malin JA, Konstantinides N, et al. Neuronal diversity and convergence in a visual system developmental atlas. Nature. 2021;589(7840):88–95. doi: 10.1038/s41586-020-2879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Farzana L, Robertson LK, Hutchinson J, Grubbs N, Shepherd MN, Mahaffey JW.. The appendage role of insect disco genes and possible implications on the evolution of the maggot larval form. Dev Biol. 2007;309(1):56–69. doi: 10.1016/j.ydbio.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Romano RA, Li H, Tummala R, Maul R, Sinha S.. Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics. 2004;83(5):821–833. doi: 10.1016/J.YGENO.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Rossi AM, Jafari S, Desplan C.. Integrated patterning programs during Drosophila development generate the diversity of neurons and control their mature properties. Annu Rev Neurosci. 2021;44:153–172. doi: 10.1146/ANNUREV-NEURO-102120-014813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, Briscoe J.. Establishing neuronal diversity in the spinal cord: a time and a place. Development. 2019;146(22). doi: 10.1242/DEV.182154/223189. [DOI] [PubMed] [Google Scholar]
- Sato M, Umetsu D, Murakami S, Yasugi T, Tabata T. DWnt4 regulates the dorsoventral specificity of retinal projections in the Drosophila melanogaster visual system. Nat Neurosci. 2006;9(1):67–75. doi: 10.1038/nn1604. [DOI] [PubMed] [Google Scholar]
- Sen SQ, Chanchani S, Southall TD, Doe CQ.. Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci. Elife. 2019;8. doi: 10.7554/ELIFE.44036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H, Fischbach KF, Rubin GM. Disconnected: a locus required for neuronal pathway formation in the visual system of Drosophila. Cell. 1987;50(7):1139–1153. doi: 10.1016/0092-8674(87)90180-2. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Hasegawa E, Nakai Y, Kaido M, Takayama R, Sato M.. Formation of neuronal circuits by interactions between neuronal populations derived from different origins in the Drosophila visual center. Cell Rep. 2016;15(3):499–509. doi: 10.1016/j.celrep.2016.03.056. [DOI] [PubMed] [Google Scholar]
- Wu M, Nern A, Williamson WR, Morimoto MM, Reiser MB, Card GM, Rubin GM, Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. Elife. 2016;5. doi: 10.7554/ELIFE.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135(8):1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Zhu H, Zhao SD, Ray A, Zhang Y, Li X.. A comprehensive temporal patterning gene network in Drosophila medulla neuroblasts revealed by single-cell RNA sequencing. Nat Commun. 2022;13(1):1–19. doi: 10.1038/s41467-022-28915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Drosophila strains and antibodies are available upon request. The raw and processed sequencing data used in this study have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE205934. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures.
Supplemental material is available at GENETICS online.