Figure 3.
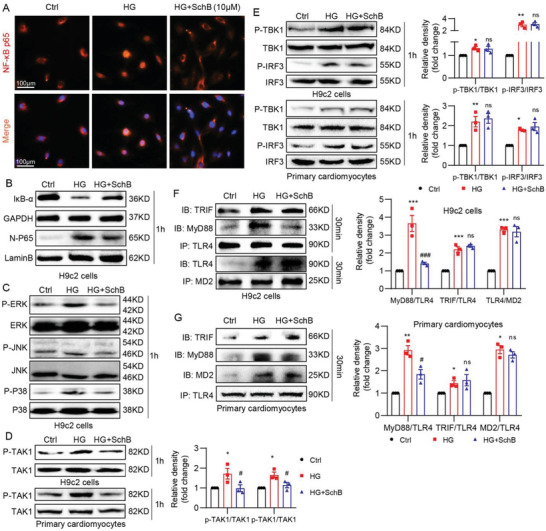
Sch B exhibits differential regulation of TLR4/TRIF and TLR4/MyD88 pathways. A) Immunofluorescence staining of H9C2 cells for p65 subunit of NF‐κB (red). Cells were pretreated with 10 × 10−6 m Sch B for 1 h and then exposed to HG for 1 h. Cells were counterstained with DAPI (blue) (scale bar = 100 µm). B) Immunoblot analysis of IκBα in total cell lysates and p65 in nuclear fractions prepared from H9C2 cells. Cells were treated as indicated for panel (A). GAPDH was used as control for total cell lysates and lamin B for nuclear proteins. C) Western blot analysis of MAPK activation in H9C2 cells. Cells were treated as in panel (B). Total cell lysates were probed for phosphorylated (p‐) and total ERK, JNK, and p38. D) Levels of MyD88 pathway protein TAK1 in H9C2 cells exposed to HG. Cells were treated as indicated for panel (B). Upper blots showing H9C2 cells and lower blots showing primary cardiomyocytes. Densitometric quantification of blots is shown in right panel. E) Analysis of TRIF pathway showing levels of TBK1 and IRF3. H9C2 (upper blots) and primary cardiomyocytes (lower blots) were treated as indicated for panel (B). Densitometric quantification is shown on right. F) Co‐immunoprecipitation analysis to examine the MD2/TLR4, TLR4/TRIF, and TLR4/MyD88 complexes in H9C2 cells. Cells were pretreated with 10 × 10−6 m Sch B for 1 h and then exposed to HG for 30 min. TLR4 was immunoprecipitated (IP) and TRIF and MyD88 were detected by immunoblotting (IB). In lower blots, MD2 was immunoprecipitated (IP) and TLR4 was detected by immunoblotting (IB). G) Co‐immunoprecipitation of MD2/TLR4, TLR4/TRIF, and TLR4/MyD88 complexes in primary cardiomyocytes. Cells were treated as indicated for panel (F). Densitometric quantification is shown in the lower panel. All data in panels (D)–(G) is shown as mean ± SEM (n = 3; *p < 0.05, **p < 0.01 compared to Ctrl and ##p < 0.01, ns = not significant compared to HG by one‐way ANOVA followed by Bonferroni's multiple comparisons test).
