Table 2. Docking Scores Obtained by Different Programs and Exponential Consensus Ranking for the Test Compounds Docked within the Active Site of α-Glucosidase and α-Amylase.
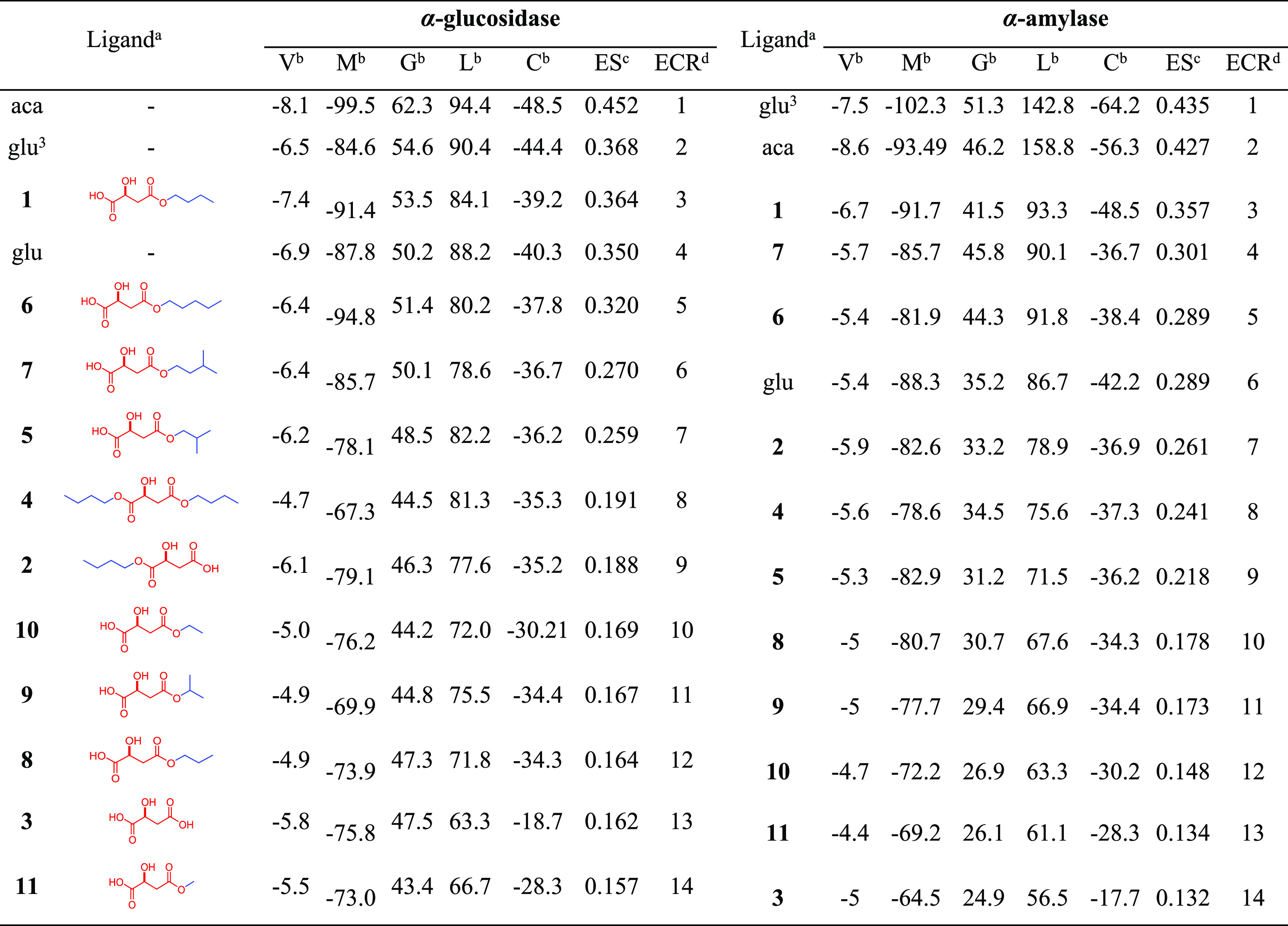
Selected ligands docked within the active site of the two test enzymes: acarbose (aca), glucose (glu), (1 → 6)-glucose trimer (glu3), (S)-4-butoxy-2-hydroxy-4-oxo-butanoic acid (1), (S)-4-butoxy-3-hydroxy-4-oxo-butanoic acid (2), (S)-malic acid (3), dibutyl (S)-malate (4), (S)-4-isobutoxy-2-hydroxy-4-oxo-butanoic acid (5), (S)-4-pentoxy-2-hydroxy-4-oxo-butanoic acid (6), (S)-4-isopentoxy-2-hydroxy-4-oxo-butanoic acid (7), (S)-4-propoxy-2-hydroxy-4-oxo-butanoic acid (8), (S)-4-isopropoxy-2-hydroxy-4-oxo-butanoic acid (9), (S)-4-ethoxy-2-hydroxy-4-oxo-butanoic acid (10), and (S)-4-methoxy-2-hydroxy-4-oxo-butanoic acid (11).
Scores obtained after molecular docking simulations with programs having different search algorithms and scoring functions: V = Vina scores, M = scores MolDock scores, G = GOLD scores, L = LibDock scores, C = CDOCKER scores.
ES = exponential scores obtained from docking scores per program through the metrics previously reported.43
ECR = exponential consensus ranking organized from ES.
