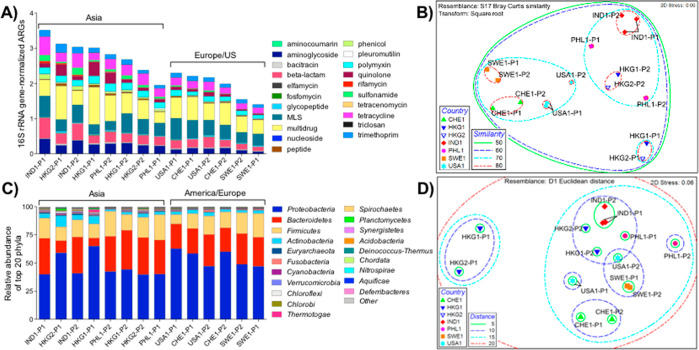
Figure 1.
A: ARG distribution and relative abundance by corresponding antibiotic categories. ARGs were annotated via MetaStorm using CARD version 1.2.1. ARG categories were assigned in-house (Supplemental Data 1). Genes corresponding to two or more categories were labeled as “multidrug”. ARG abundances were normalized via MetaStorm to 16S rRNA gene abundances. B: Nonmetric multidimensional scaling (NMDS) ordination of WWTPs according to the ARG-based Bray–Curtis dissimilarities. Ellipses enclose sites of noted similarities, ranging from 0 to 100 (perfect similarity); thus, ellipses with a similarity of 80 represent sites with highest similarity. C: Relative abundance of the top 20 bacterial phyla in WWTP influents. Genus-level annotations were done via MG-RAST using the RefSeq database. D: NMDS ordination of WWTPs according to Euclidian distances between rlog-transformed reads to bacterial genera. Ellipses enclose sites of noted distances, where the shortest distance denotes the highest resemblance, and the longest distance denotes the least resemblance among the sites. Sample names refer to countries (IND: India, PHL: Philippines, USA: United States, CHE: Switzerland, HKG: Hong Kong, SWE: Sweden) along with the visit number (1,2) and WWTP plant number (P).