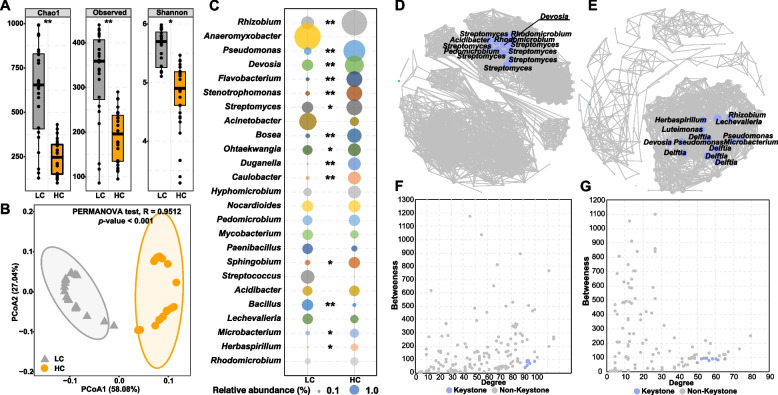
Fig. 1.
The boxplots show the bacterial alpha diversity indices in the root endosphere at the low contaminated (LC) and the high contaminated (HC) sites (A). The PCoA plot of beta similarities was measured as Bray–Curtis distances for the bacterial community (B). Comparison of the different genera distribution between LC and HC sites (C). * and ** indicate significant differences between the LC and HC at p < 0.05 and p < 0.01, respectively. Co-occurrence network analysis showing the biological interactions in the LC (D) and HC (E). Edges are shown only strong (Spearman correlation >|0.6|) and significant (p < 0.05) connections. The size of the nodes is proportional to the number of connections to it. The scatter plot shows criteria of selecting for the keystone taxa in the LC (F) and HC (G). The bacterial community was analyzed based on 5 replicate root samples from each sampling site (a total of 20 root samples)