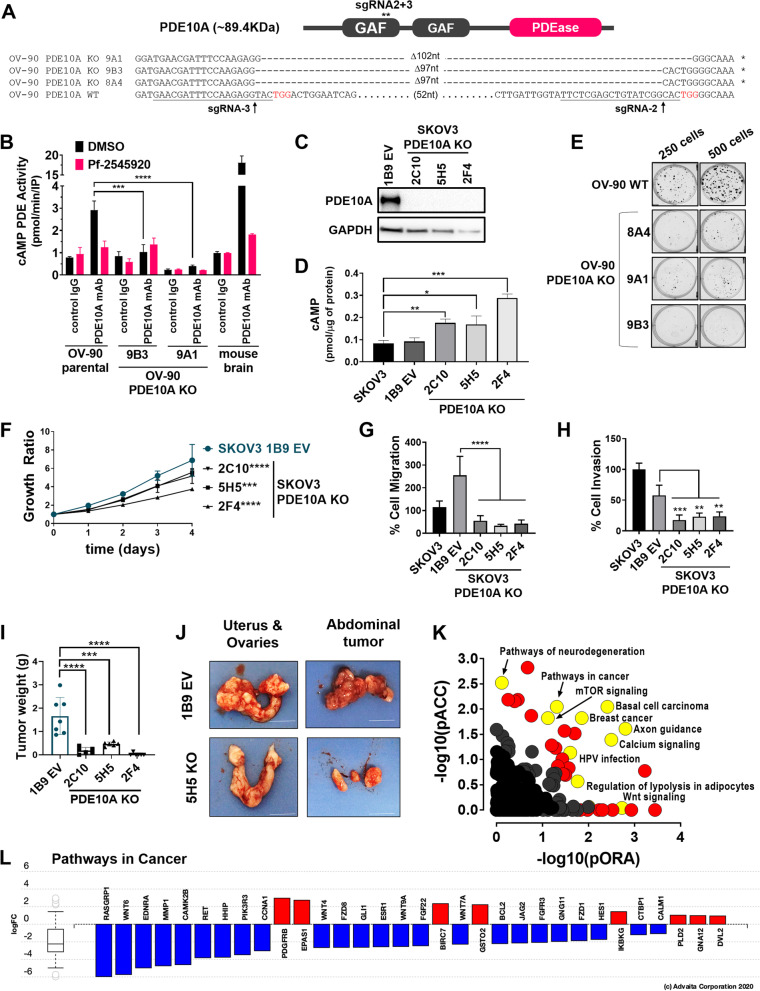
Fig. 2.
Antineoplastic effects of PDE10A gene knockout (KO) in ovarian cancer cells. A On top, schematic representation of PDE10A protein showing two cyclic nucleotide binding GAF domains at the N-terminus, and a phosphodiesterase catalytic domain (PDEase) at the C-terminus. On bottom, the genomic locus of PDE10A exon 7 was targeted by two CRISPR/Cas9 guide RNAs (sgRNAs). The predicted Cas9 cleavage sites are located 97 bp apart in exon 7. Successful deletion of the intended DNA fragment in OV-90 PDE10A KO clones in comparison to wild-type (WT) parental cells was confirmed by PCR followed by Sanger DNA sequencing. B PDE10A enzymatic activity in OV-90 parental and PDE10A KO clones was evaluated by immunoprecipitation of PDE10A followed by a cAMP-phosphodiesterase activity assay. Mouse brain tissue lysate was used as positive control. Pf-2545920 treatment was used to confirm that the measured activity was PDE10A-specific. C Confirmation of successful PDE10A KO in SKOV3 clones by western blotting. GAPDH was used as the loading control. D Baseline cAMP levels detected by ELISA in SKOV3 parental and PDE10A KO cells. Statistical significance was also observed when comparing KO clones to EV 1B9 control cells. Error bars, ± SD, ***p < 0.001, **p < 0.01, *p < 0.05 (student’s t-test). E Colony formation assays in OV-90 WT and PDE10A KO clones after 12 days of growth. F Proliferation of SKOV3 PDE10A KO clones. Growth ratios calculated by normalizing to day-0 seeding densities. p-values represent statistical significance on day 4. Error bars, ± SD, ***p < 0.001, ****p < 0.0001 (2-way ANOVA). G-H Migration (G) and invasion (H) of SKOV3 cells and PDE10A KO clones in the Boyden chamber assay over 24 h. Error bars, ± SD, ****p < 0.0001 (Ordinary one-way ANOVA). I Total tumor burden of athymic nude mice injected i.p. with SKOV3 PDE10A KO clones (n = 6 per group) or SKOV3 empty vector (EV 1B9) control cells (n = 7). Tumor burden included total tumor weights from injection site, uterus and ovary, and mesentery or abdominal tumors. ***p < 0.001, ****p < 0.0001 (Ordinary one-way ANOVA). J Representative images of uterus and ovaries and abdominal tumors from control and PDE10A KO nude mice groups. Scale bar = 1 cm. K iPathway Guide impact analysis of RNAseq results of DESeq2 differentially expressed genes comparing SKOV3 PDE10A KO clones (5H5 and 2F4) vs. SKOV3 parental and empty vector (1B9) control cells. In this comparison, 833 differentially expressed (DE) genes were identified out of a total of 14,195 genes with measured expression using thresholds of log2-fold change ± 0.6 and adjusted p-value < 0.05. Each pathway is represented as a single dot, with significant pathways shown in red and yellow, and non-significant in black. L Log2-fold change expression for “Pathways in Cancer” gene set. Upregulated genes are depicted in red and downregulated genes are depicted in blue