Figure 1. A base editor screen to investigate regulatory relationships affecting protein levels.
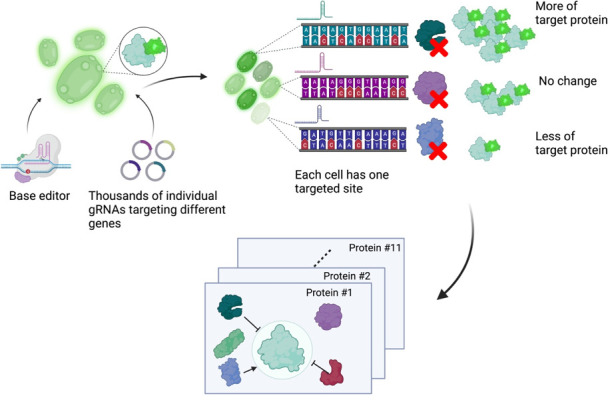
Left: a library of thousands of guide RNA plasmids (gRNA, circles with sections in different colours) targeting genes across the genome are introduced, along with the base editor, into an engineered yeast strain containing the genetic code for a protein of interest fused to the fluorescent protein GFP. Centre: each cell has a different target gene disrupted by the base editor (shown as DNA sequences in different colours), resulting in a specific protein (the green, purple and blue shapes next to the sequences) not being produced in each cell. Right: after editing, each cell will have different levels of fluorescence depending on which gene was disrupted in it. The top cell has higher fluorescence than an unedited cell because the disrupted gene in it codes for a protein that normally lowers the levels of the protein of interest. The centre cell has the same fluorescence levels as an unedited cell because the disrupted gene in it codes for a protein that does not regulate the levels of the protein of interest. The bottom cell has low levels of fluorescence because the disrupted gene in it codes for a protein that normally increases the levels of the protein of interest. The cells are then sorted depending on their fluorescence levels, and the identity of the gene mutated in each cell is determined by sequencing the gRNA. Bottom: this pooled screening approach allows scientists to define regulatory networks that determine the abundance of each target protein and reveals general properties of networks governing protein expression.
Figure created using BioRender.com
