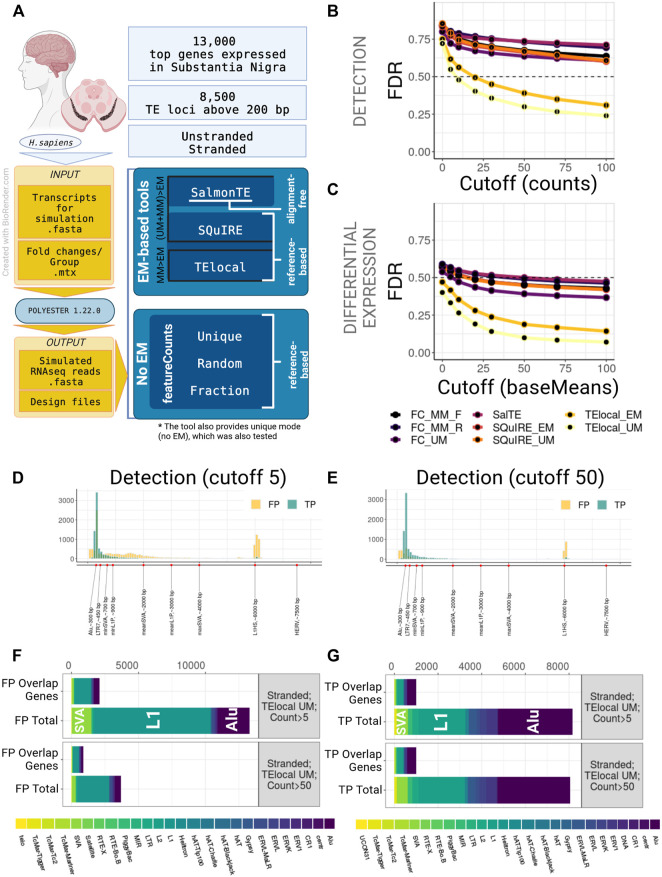
FIGURE 1.
Benchmarking of TE quantification tools on human model, stranded experiment. (A) A general overview of the simulation setup and the strategies used in benchmarking. For human simulation 2,000 TEs over 200 bp long and top 13,000 genes expressed in substantia nigra were simulated in stranded and unstranded experiments using Polyester 1.22.0. The resulting simulated sequencing data was processed using 3 EM-based tools (both in EM and no EM modes, where permissible) and 3 modes of featureCounts. (B) TE Detection FDR for different detection cutoffs using the tested tools. TElocal in unique mode (TElocal_UM) outperformed other strategies closely followed by TElocal in EM mode (TElocal_MM), however even with higher cutoffs FDR reached 26%. (C) TE Differential Expression detection FDR for different expression cutoffs. (D,E) Length distribution of True Positive (TP) and False Positive (FP) hits for TElocal in UM mode at detection cutoff 5 (D) and 50 (E). (F) Family Composition of total FP hits (Total) and FP hits overlapping the simulated expressed genes (Overlap Genes) for TElocal_UM at detection cutoffs 5 and 50. Only a minority of the FPs at both cutoffs can be explained by misattribution of the genic reads (2091/13301 for cutoff = 5 and 887/3705 for cutoff = 50). (G) Family Composition of TP hits categorized by total and overlap genes. FC_MM_F, featureCounts using multimappers in “fraction” mode; FC_MM_R, featureCounts using multimappers in “random” mode; FC_UM, featureCounts using unique mappers only; SalTE, SalmonTE; SQuIRE_EM, SQuIRE in EM mode; SQuIRE_UM, SQuIRE in unique mode; TElocl_EM, TElocal in EM mode; TElocal_UM, TElocal in unique mode.