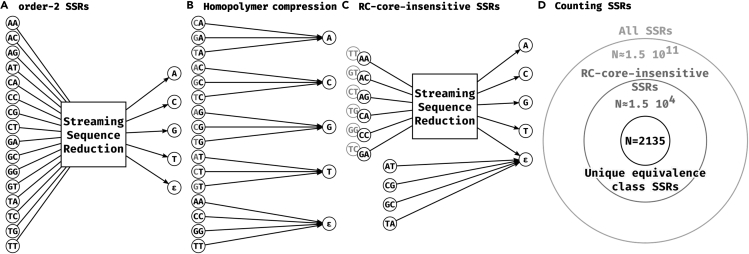
Figure 1.
Representing and counting streaming sequence reductions
(A) General representation of an order-2 streaming sequence reduction as a mapping of 16 input dinucleotides, to the 4 nucleotide outputs and the empty character ε.
(B) Homopolymer compression is an order-2 SSR. All dinucleotides except those that contain the same nucleotide twice map to the second nucleotide of the pair. The 4 dinucleotides that are the two same nucleotides map to the empty character ε.
(C) Our RC-core-insensitive order-2 SSRs are mappings of the 6 representative dinucleotide inputs to the 4 nucleotide outputs and the empty character ε. The 4 dinucleotides that are their own reverse complement are always mapped to ε. The remaining 6 dinucleotides are mapped to the complement of the mapped output of the reverse complement dinucleotide input. For example, if AA is mapped to C, then TT (the reverse complement of AA) will be mapped to G (the complement of C).
(D) Number of possible SSR mappings under the different restrictions presented in the main text. All mappings from 16 dinucleotide inputs to 5 outputs (as in panel A) are represented by the outermost circle. All RC-core-insensitive mappings (as in panel C) are represented by the medium circle. All RC-core-insensitive mappings with only one representative of each equivalence class are represented by the innermost circle.