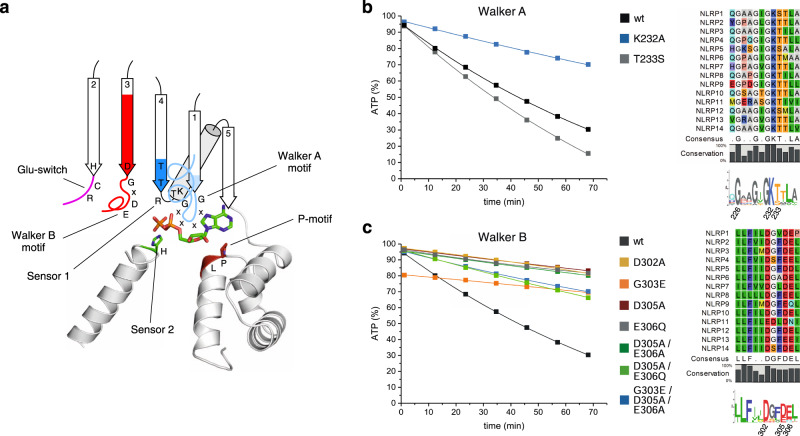
Fig. 3. Mutational analysis of Walker A/B nucleotide-binding motifs in NLRP3.
a A cartoon of the nucleotide-binding site of NLRP3 including ADP is shown based on the structure of inactive NLRP3 (PDB: 7PZC). The Walker A and B motifs as well as the Glu-switch, Sensor 1, Sensor 2 and the P-motif are indicated. b MBP-NLRP3 wild type and Walker A motif point-mutants were analyzed by RP-HPLC. Sequence alignments and sequence logos of the labeled motifs and residues are shown for human NLRP1-14. Protein samples (3 µM) were incubated at 25 °C in the presence of 100 µM ATP and 5 mM MgCl2. The amount of ATP (%) is shown on the y-axis and the time (min) on the x-axis. The amount of ATP was determined in 10 min intervals using RP-HPLC, integrated and normalized to 100% nucleotide. c MBP-NLRP3 wild type and Walker B motif point-mutants were analyzed by RP-HPLC using the same set-up as in b. All mutant significantly reduced the hydrolysis activity.