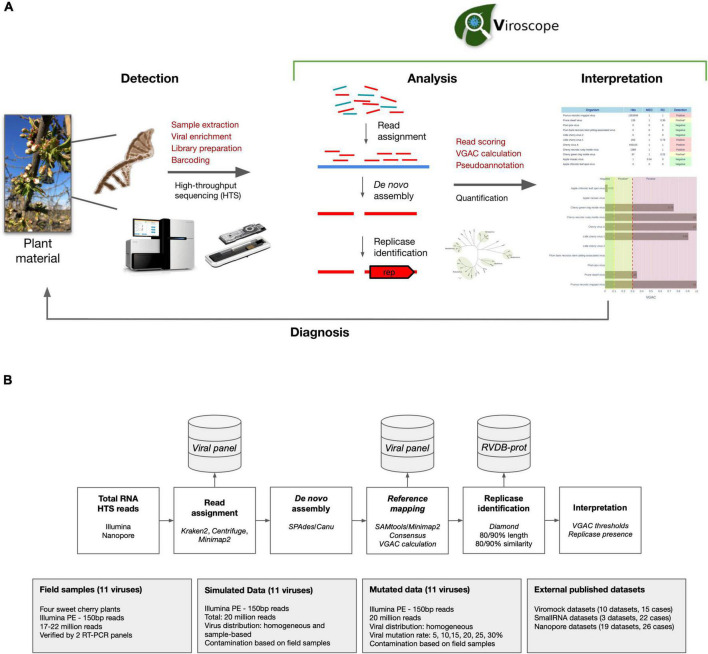
FIGURE 1.
Overview of Viroscope, the data analysis pipeline and experimental validation strategy. (A) Overview of Viroscope. Viroscope is a total RNA HTS data analysis pipeline that enables accurate viral detection by performing read assignment, de novo assembly with reference-based mapping and pseudo-annotation to obtain VGAC metrics (Viral Genome Assembly Coverage) and identification of viral replicases. These metrics inform the interpretation of viral presence from HTS reads to provide accurate diagnosis, contributing to the implementation of HTS for plant viral diagnosis in real-world applications. (B) The Viroscope pipeline and experimental validation strategy. Viroscope performs read assignment from total RNA HTS (Illumina or Nanopore) reads using 3 read assignment software against a curated database of target viruses. Then, mapped reads are collected and used for de novo assembly either using SPAdes or Canu (for Illumina and Nanopore reads, respectively). Assembled contigs are used to perform reference-based mapping to obtain a consensus of mapped contigs to calculate VGAC. Then, the pipeline searches for the presence of replicases using Diamond using the RVDB-prot database. Finally these metrics are used for interpretation of viral presence according to specific cutoffs for diagnosis. Four experimental sets were used to validate the pipeline, sweet cherry field samples, a simulated dataset, a mutation dataset and external published datasets.