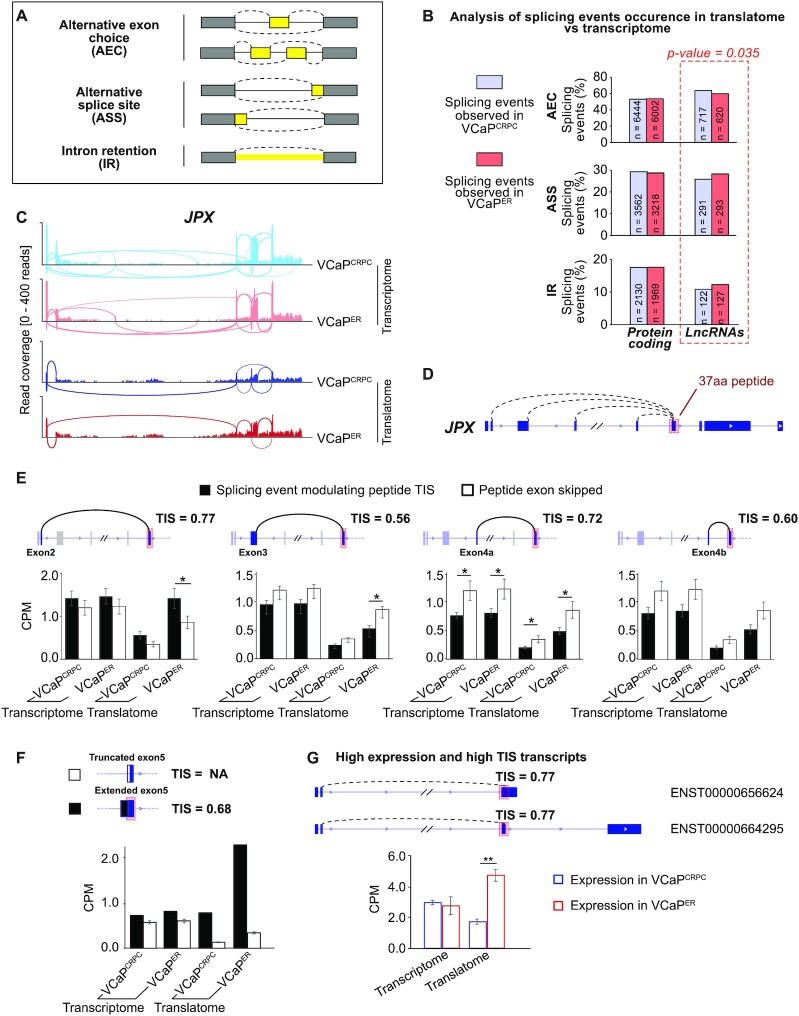
Figure 5.
Alternative splicing of lncRNAs promotes putative coding isoforms in VCaPER. (A) Schematic of alternative splicing event types. (B) Analysis of alternative splicing events with rMATS (53,54) to compare splicing events occuring in the translatome to those in the total transcriptome. Bargraphs show percentage of alternative events detected and compared through chi-squared test. Number of events are indicated. (C) Read density histograms of the JPX locus for transcriptomes and translatomes of VCaPCRPC or VCaPER. (D) Schematics of the JPX locus with exons (boxes) and introns (lines) indicated. Black dashed curves indicate alternative splicing events modifying the putative peptide's (exon 5) TIS. (E) Schematics of JPX exons 1 to 5 and of alternative splicing events changing the putative peptide's TIS (top). Predicted TIS scores are indicated. Quantification of corresponding transcripts in VCaPCRPC and VCaPER translatomes and transcriptomes (black) and of transcripts skipping exon 5 (white) (bottom). Dark blue boxes: included exons, pastel blue: potentially included exons, grey: excluded exons. Of note, exons 4a and 4b are mutually exclusive. (F) Schematic of JPX exon 5 and quantification of transcripts with alternative splice site choice for exon 5. Extended (white) and truncated (black) exon 5 are shown with corresponding TIS scores. (G) Schematic of the JPX locus and quantification of high-TIS score (0.77) transcripts in VCaPCRPC (blue) and VCaPER (red) translatomes and transcriptomes. Red boxes indicate putative peptide-coding sequences. Error bars indicate standard error of the mean, SEM. *P-value < 0.05; **P-value < 0.01.