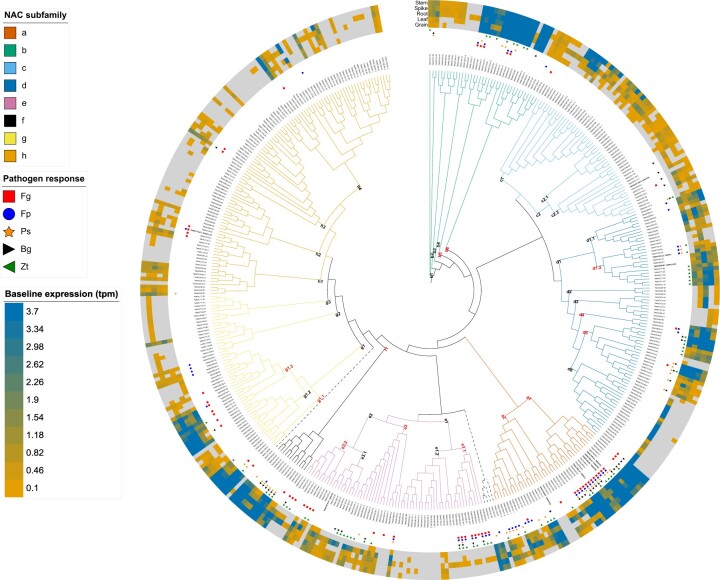
Fig. 4.
Consensus maximum likelihood tree of TaNAC proteins with their corresponding healthy tissue and disease-responsive gene expression profiles indicated (https://itol.embl.de/tree/372282299294211613865063). Branch lines are colored according to NAC subfamily (unclassified branches and clades are indicated with a dashed line). Within a subfamily, TaNAC subclades are coded at the node based on the subfamily letter and a specific number. For example, “e1” is the first subclade of subfamily “e,” and “e1.1” and “e1.2” are two distinct subclades within “e1” subclade. Red font indicates subclades enriched with pathogen-responsive TaNACs. TaNACs highlighted in bold are functionally characterized in pathogen defense (Table 3). The colored symbols denote the TaNACs that are responsive (at the transcriptome level) to pathogens. Pathogen abbreviations: Fg, Fusarium graminearum; Fp, Fusarium pseudograminearum; Ps, Puccinia striiformis; Bg, Blumeria graminis; Zt, Zymoseptoria tritici. TaNAC expression within healthy tissues (grain, leaf, root, spike, and stem) is represented by a color scale that represents baseline expression in tpm values, and the color gradient maximum is set to 3.7 tpm.