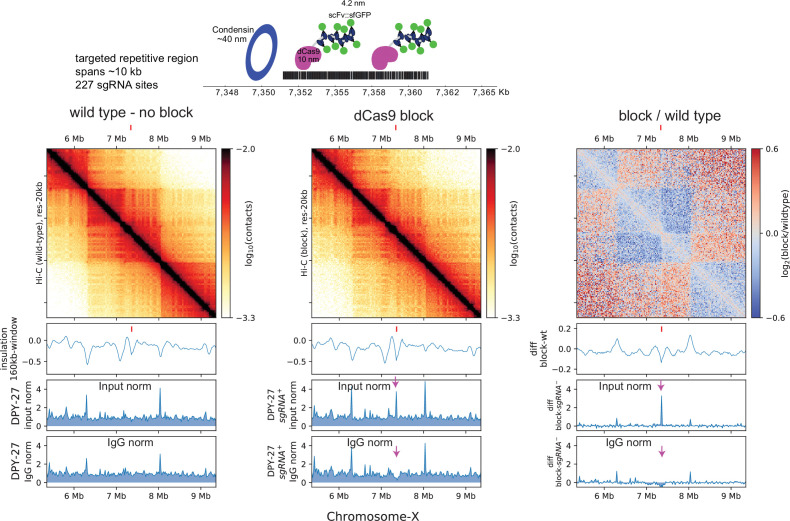
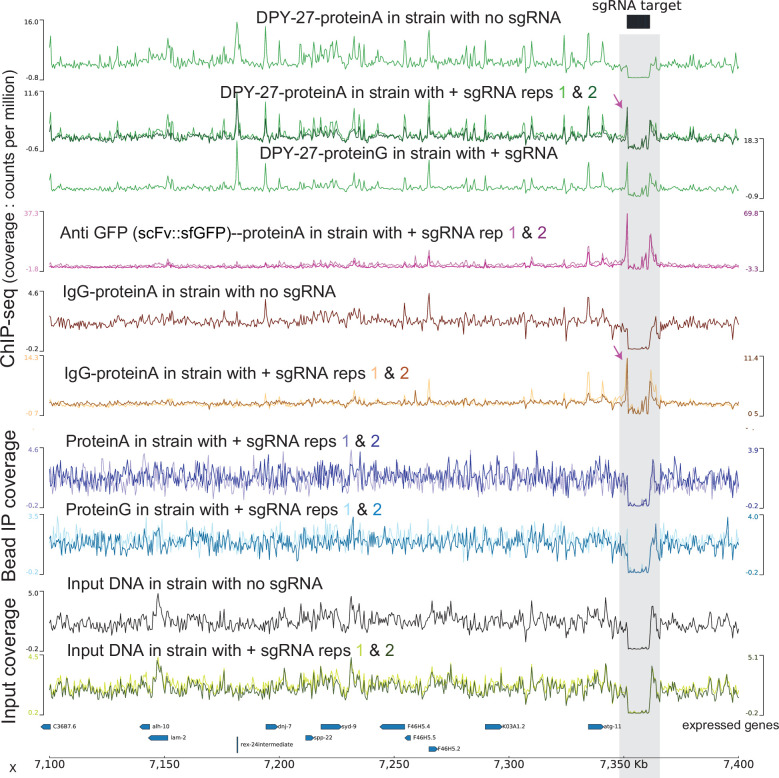
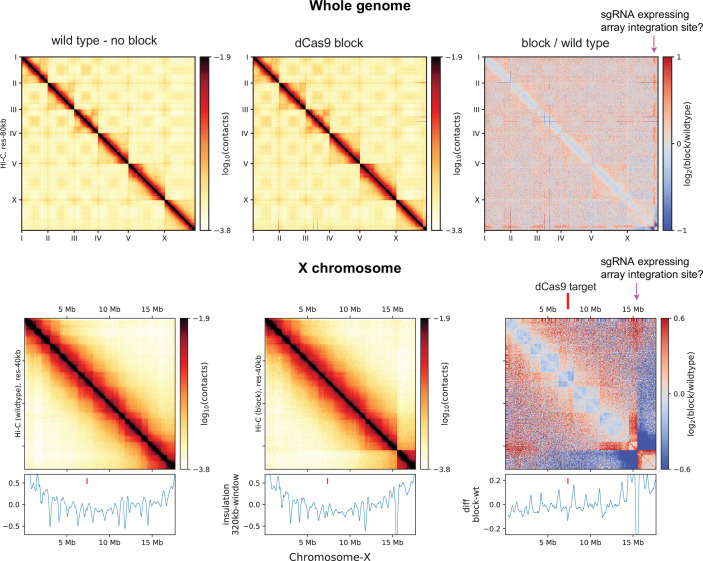
Figure 4. dCas9 block fail to sufficiently recapitulate rex-like boundary on the X-chromosome.
Top: schematic depicting the multi-protein block and the approximate size of the components utilized to prevent condensin from translocating linearly along chromatin. Bottom: Hi-C of wildtype and block strain expressing all components of dCas9-SunTag and single guide RNA (sgRNA). The tick marks point to the block target. The below are ChIP-seq data in dCas9-SunTag expressing strain with (first panel) and without (middle panel) sgRNA. The two ChIP-seq tracks show normalization by input and IgG. Arrows pointing down to the DPY-27 ChIp signal that is apparent when data is normalized to input but not to IgG.