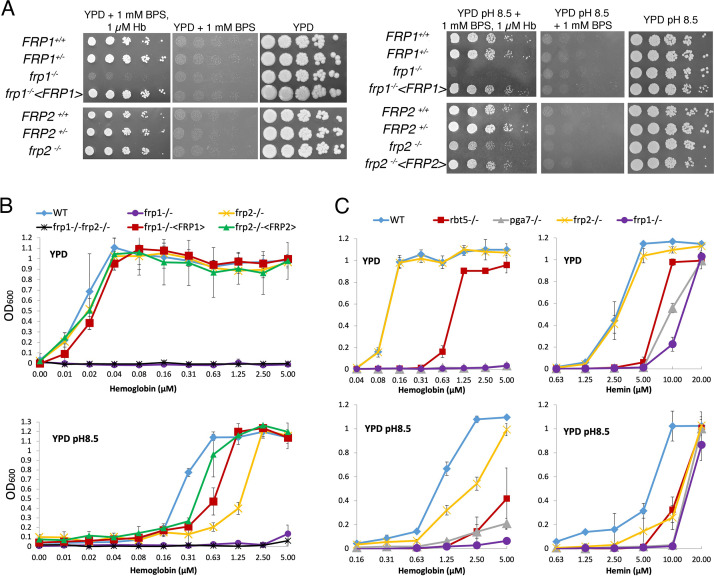
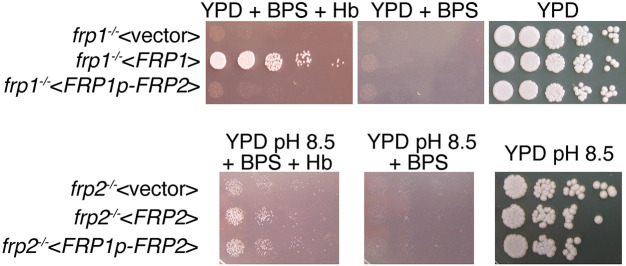
Figure 1. FRP1 is essential for hemoglobin-iron acquisition, whereas FRP2 contributes to growth on hemoglobin at alkaline pH.
(A) Fivefold dilutions of cultures of the indicated strains were spotted on YPD or YPD pH 8.5, with the indicated supplements, and incubated for 3 days (Hb and BPS plates) or 2 days (YPD plates) at 30°C. WT = KC2, FRP1+/-=KC859, frp1-/-=KC870, frp1-/-<FRP1 >= KC1024, FRP2+/-=KC901, frp2-/-=KC912, frp2-/-<FRP2 >= KC1379. (B) The strains with indicated genotypes in the ccc2-/- background were grown in triplicates in YPD or YPD pH 8.5 media supplemented with 1 mM ferrozine and the indicated amounts of hemoglobin, and incubated at 30°C for 3 days. Each result is the average of three cultures. Standard deviations are indicated by vertical bars. WT = KC811, frp1-/-=KC1146, frp2-/-=KC1414, frp1-/- frp2-/-=KC1412, frp1-/-<FRP1 >= KC1146, frp2-/-<FRP2>=KC1411. (C) The frp1-/- and frp2-/- heme-iron utilization phenotype was compared to that of the CFEM protein mutants rbt5-/- and pga7-/-. The strains were grown in YPD or YPD pH 8.5, with 1 mM ferrozine and the indicated concentrations of hemoglobin or hemin, and grown and measured as in B. Wild type = KC68, rbt5-/-=KC139, pga7-/-=KC485, frp1-/-=KC923, frp2-/-=KC913. All strains in B and C carry a deletion of the CCC2 gene, which causes a defect in high-affinity iron import and prevents growth in the presence of ferrozine.