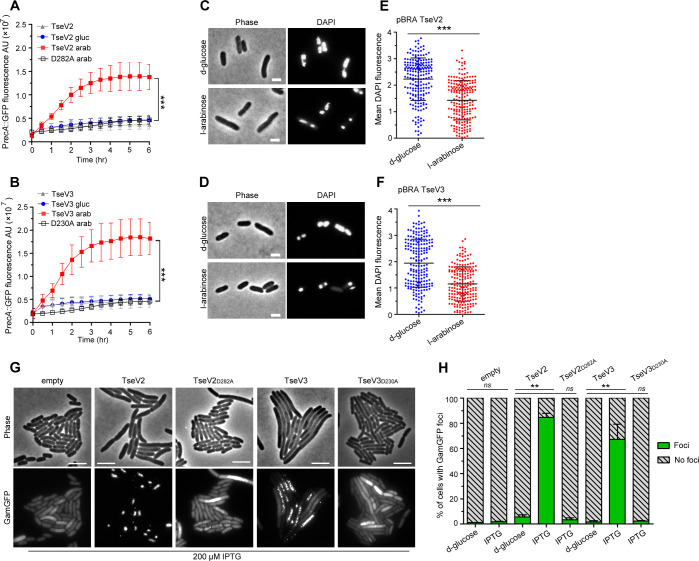
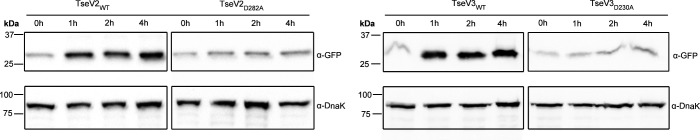
Figure 4. TseV2 and TseV3 induce DNA double-strand breaks.
Activation of the SOS response was analyzed using E. coli cells harboring the reporter plasmid pSC101-PrecA::GFP and pBRA TseV2 (A) or pBRA TseV3 (B), which were grown in AB defined media with d-glucose or l-arabinose. Data is the mean ± standard deviation (SD) of three independent experiments. ***p < 0.001 (Student’s t-test). Bright-field and DAPI images of E. coli cells carrying pBRA TseV2 (C) or pBRA TseV3 (D) grown in the presence of d-glucose (repressed) or l-arabinose (induced). Results are representative images of three independent experiments. (E, F) Quantification of the mean 4′,6-diamidino-2-phenylindole (DAPI) fluorescence per cell of 200 cells. Data correspond to the mean ± SD of a representative experiment. Scale bar 2 μm. ***p < 0.001 (Student’s t-test). (G) Representative bright-field and GFP images of E. coli coexpressing GamGFP and pEXT20 TseV2 or pEXT20 TseV3. Double-strand breaks appear as foci of GamGFP. Images are representatives of three independent experiments. Scale bar: 5 μm. (H) Quantification of the GamGFP foci shown in (G). Data are shown as the mean ± SD of the three independent experiments. **p < 0.01 (Student’s t-test).