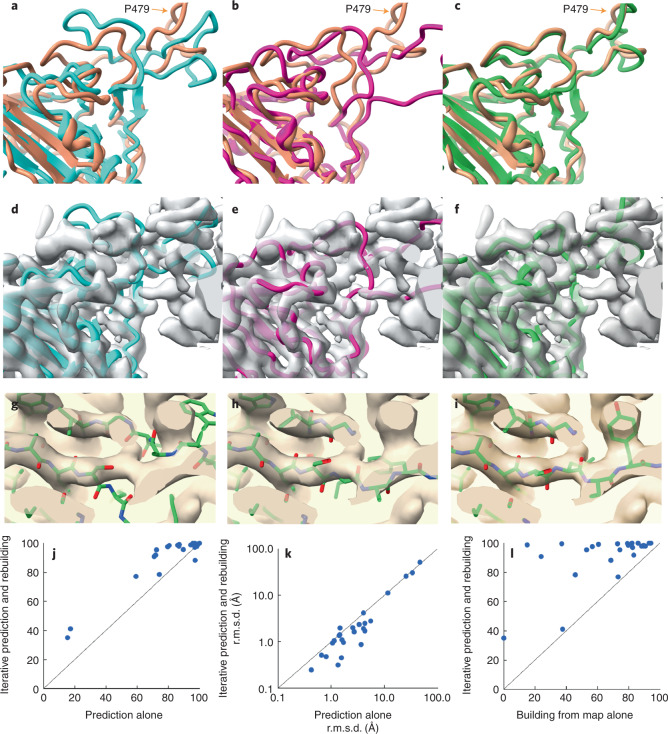
Fig. 1. Iterative AlphaFold prediction and model rebuilding using density maps.
a, Comparison of AlphaFold model of SARS-CoV-2 spike protein RBD (blue) with deposited model16 (PDB entry 7MLZ, brown). The position of P479 is indicated. b, Comparison of model in a rebuilt using density map (in purple) with deposited model (brown). c, AlphaFold model obtained using density map and four cycles of iteration including rebuilt models as templates (green), compared with deposited model (brown). d–f, Models as in a–c, superimposed on the map used for rebuilding (EMDB entry 23914 (ref. 26), automatically sharpened as described (Methods)). g–i, Details of iterative rebuilding of the 2AG3 Fab heavy chain17 (PDB entry 7MJS chain H) using cryo-EM data from EMDB entry 23883 at a resolution of 3.0 Å. g, AlphaFold prediction superimposed on density map. h, AlphaFold prediction as in g, but after one cycle of iterative rebuilding. i, As in h, but after four cycles of iterative rebuilding. j, Accuracy of models obtained with AlphaFold alone (abscissa) and obtained with iterative AlphaFold prediction and rebuilding with density (ordinate) for one chain from each of 25 structures from the PDB and EMDB. Accuracy is assessed as the percentage of of Cα atoms in the deposited model matched within 3 Å by a Cα atom in the superimposed AlphaFold model. k, Accuracy of models shown in j, assessed on the basis of r.m.s.d. of matching Cα atoms and shown on a log scale. Abscissa is r.m.s.d. for models obtained with AlphaFold alone and ordinate is for models obtained with iterative AlphaFold prediction and rebuilding with density. l, Accuracy of models assessed as in j by the percentage of of Cα atoms in the deposited model matched within 3 Å by a Cα atom in the superimposed model, obtained with direct model building using the corresponding density maps using the Phenix tool map_to_model (abscissa) compared with those obtained with iterative AlphaFold prediction (ordinate).