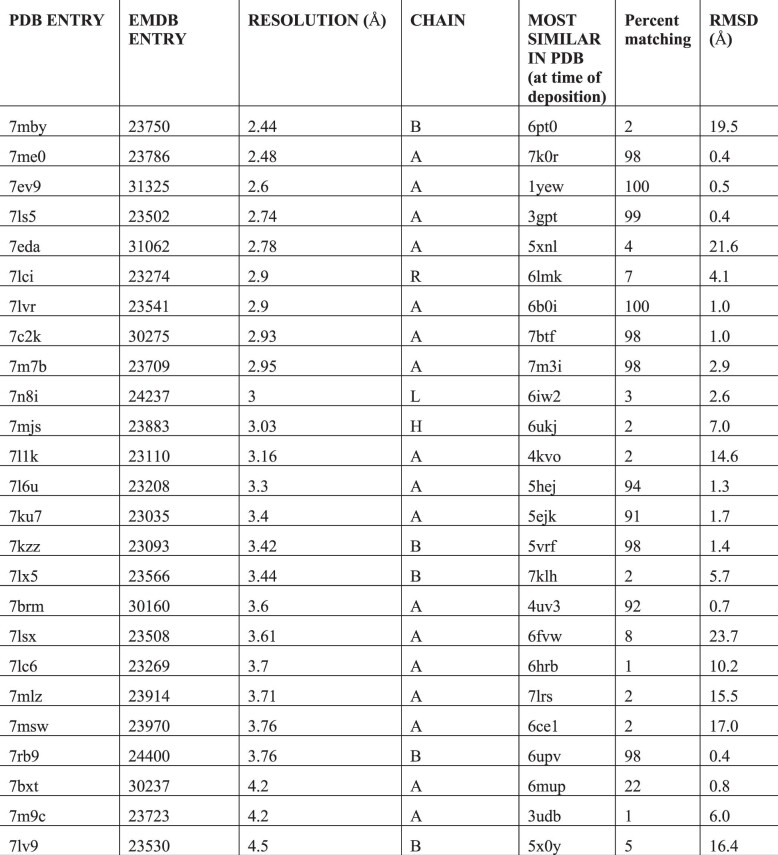
Extended Data Table 1.
Structures and maps used
List of structures and maps used in Figs. 1 and 3. The most similar entries at time of deposition were obtained using the RCSB PDB ‘Find similar proteins by 3D structure tool’ and choosing the highest-scoring entry that was deposited earlier than the target structure. The percentage of residues matching and rmsd were obtained using the Phenix superpose_pdbs tool using only Ca atoms from the target and noting the number of residues in the target, the number of superposed residues, and the final rmsd of matching Ca atoms.