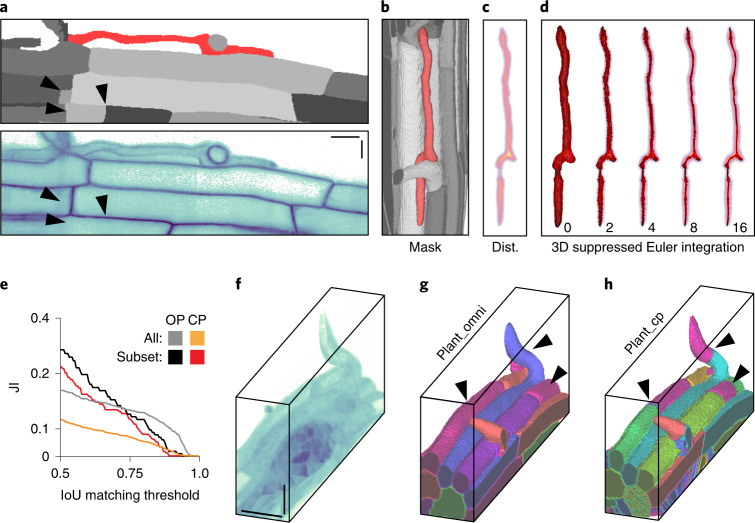
Fig. 5. Omnipose can be applied to 3D datasets.
a, Volume slice in the A. thaliana 3D training set. Black arrows denote over-segmentation in the ground truth labels. Red corresponds to the cell in b–d. b, Selected cell in context of ground-truth data. c, Ground-truth distance field for selected cell in b. d, Steps of the suppressed Euler integration for selected cell in b. Boundary pixels shown, colored by overall displacement (dark to light red). e, Performance of Omnipose (OP, plant_omni) and Cellpose (CP, plant_cp) on the full test dataset (n = 604) and on a subset without excluded regions (Methods) depicted in f–h (n = 73). f–h, Ground-truth masks (f) and segmentation results of A. thaliana using Omnipose (g) and Cellpose (h). Scale bars, 20 μM.