Figure 7.
Motifs and kinome analyses in the phosphoproteomics datasets
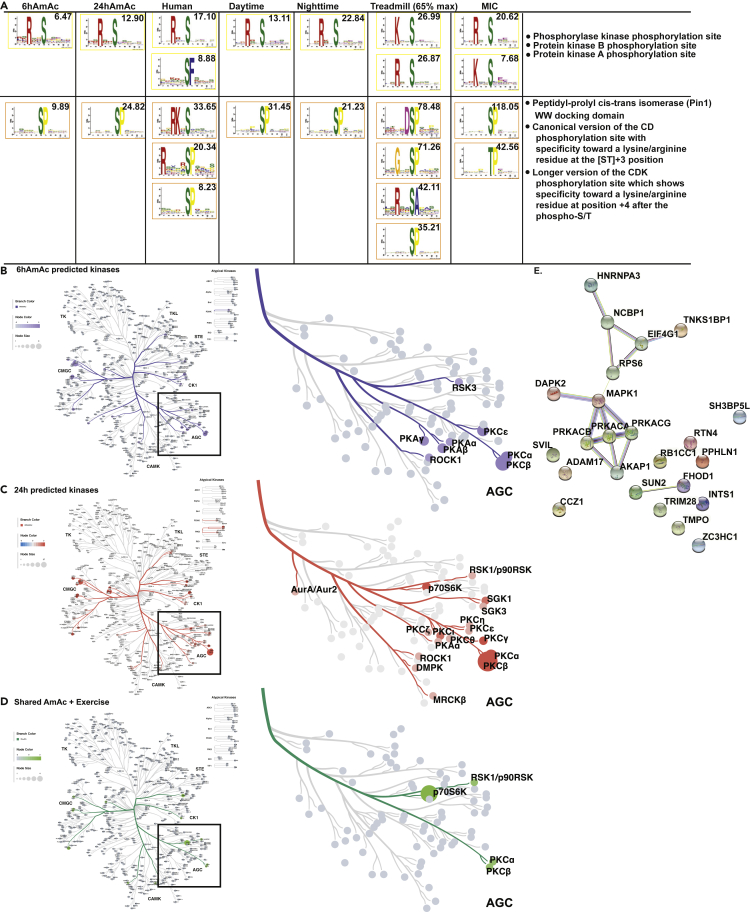
(A) Motifs and predicted kinases identified in differentially phosphorylated phosphosites (DPPS) from myotubes treated with 6 and 24h of 0mM ammonium acetate (AmAc) and skeletal muscle from mice and human exercise models. Scores shown in each panel indicates best fit with a known motif from the Eukaryotic Linear Motif database.
(B–D) Predicted kinases using NetworKIN, NetPhorest, and weighted CORAL kinome trees for DPPS in the 6hAmAc and 24hAmAc datasets and DPPS shared between any hyperammonemic (6hAmAc, 24hAmAc) dataset and any mouse or human exercise datasets. Enlarged kinome tree subsets show predicted protein kinases from the A, G and C (AGC) family in the respective datasets. Weighting is based on NetworKIN enrichment score.
(E) STRING protein-protein interaction network shows PKA, MAPK1, and their known interactions with DEpP shared between the hyperammonemia and exercise datasets. All myotube experiments were done in n = 3 biological replicates (one 24hAmAc replicate was removed from downstream analyses because of outlier status).