Table 1.
Summary of chemical modifications of Cas9 crRNAs
| Modification | Prosa | Consb | Reference | Key result | Cells | Assays |
|---|---|---|---|---|---|---|
|
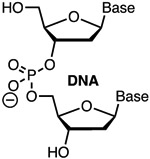
Comav Inexpn Simple | Modstb | Gagnon32 | 18 5′-terminal DNA tolerated in crRNA | In vitro | In vitro |
| Xue, Langer, Anderson33 | 10 5′-terminal DNA in crRNAs abolish off-target activity in Hepa1-6 and HEK293 cells. Optimized crRNA has 8 5′- and 16 3′-DNA | HEK293T cells constitutively expressing GFP and Cas9 | GFP fluorescence, GUIDE-seq | |||
|
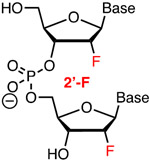
Comav Affinity | Modstb | Bennett, Cleveland42 | Seven 5′-terminal 2′-F, part of optimized crRNA that reduced off-target activity | HEK293T, tracrRNA and Cas9 plasmid | Surveyor |
| Anderson58 | Structure-guided 2′-F modification of spacer is tolerated in highly modified sgRNA | In vivo (mouse liver) | TIDE | |||
| Khvorova, Watts, Sontheimer59 | Structure-guided modification of >80% as 2′-OMe or 2′-F in crRNA/tracrRNA increases activity | HEK293T cells constitutively expressing mCherry | mCherry fluorescence, TIDE | |||
|
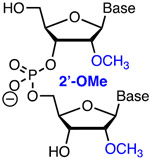
Comav Inexpn Affinity Metstab | Steric | Bruhn, Porteus41 | Three 3′- and 5′-terminal 2′-OMe slightly increased the activity of sgRNA | K562, CD34+ and T cells, Cas9 plasmid | T7EI, TIDE |
| Anderson58 | Structure-guided 2′-OMe modification is tolerated in invariable region (but not in spacer) of highly modified sgRNA | In vivo (mouse liver) | TIDE | |||
| Khvorova, Watts, Sontheimer59 | Structure-guided modification of >80% as 2′-OMe or 2′-F in crRNA/tracrRNA increases activity | HEK293T cells constitutively expressing mCherry | mCherry fluorescence, TIDE | |||
|
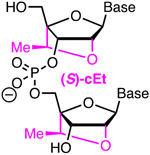
Affinity Metstab | Synth Costly Steric | Bennett, Cleveland42 | Five (S)-cEt in shortened (12-nt) repeat region part of optimized crRNA that reduced off-target activity | HEK293T, tracrRNA and Cas9 plasmid | Surveyor |
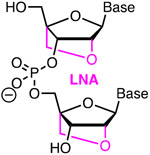
|
Amidite Affinity Metstab | Toxic Costly Steric | Hubbard43 | Up to six LNA are tolerated but less effective than BNANC in reducing off-target activity | U2OS and HeLa cells constitutively expressing Cas9 | T7EI, NGS |
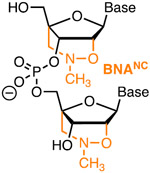
|
Affinity | Synth Costly Steric | Hubbard43 | Up to six BNANC slightly reduce Cas9 on-target activity while broadly reducing off-target activity | U2OS and HeLa cells constitutively expressing Cas9 | T7EI, NGS |
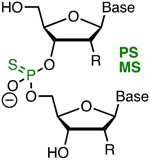
|
Comav Metstab Inexpn Simple Phrmkin | Lowaff Toxic Diast | Bennett, Cleveland42 | Full PS (R = OH) modification of crRNA increased activity ~four-fold | HEK293T, tracrRNA and Cas9 plasmid | Surveyor |
| Bruhn, Porteus41 | Three 3′- and 5′-terminal MS (R = OMe) increased the activity of sgRNA | K562, CD34+ and T cells, Cas9 plasmid | T7EI, TIDE | |||
| Anderson58 | Structure-guided PS modification is tolerated at the 3′- and 5′-ends of highly modified sgRNA | In vivo (mouse liver) | TIDE | |||
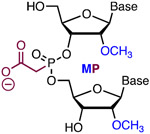
|
Simple Metstab | Costly | Bruhn50 | MP of phosphates 4, 5 and 7; 9-12; and 14 and 16 improved specificity >10-fold | K562, iPS, and CD34+ cells | NGS |
|
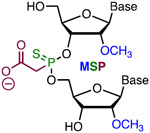
Simple Metstab | Costly | Bruhn, Porteus41 | Three 3′- and 5′-terminal MSP strongly increased the activity of sgRNA | K562, CD34+ and T cells, Cas9 plasmid | T7EI, TIDE |
|
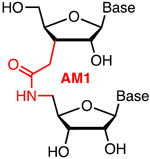
Metstab | Synth Costly | Rozners51 | Individual AM1 were well tolerated in the PAM-distal region of crRNAs | HEK293 cells constitutively expressing Cas9 | T7EI |
Abbreviations for Pros: Affinity – enhanced affinity for complementary nucleic acids, Amidite – amidite is commercially available, Comav – commercially available as an RNA modification, Inexpn – inexpensive to manufacture, Metstab – enhanced metabolic stability, Phrmkin – improved pharmacokinetics, Simple – simple synthesis.
Abbreviations for Cons: Costly – high cost, Diast – diastereomeric mixtures lead to heterogeneous compounds, Lowaff – reduced affinity for complementary nucleic acids, Modstb – modest metabolic stability, Steric – steric bulk may interfere with RNA-protein interactions, Synth – multistep synthesis of monomers required, Toxic – potential toxicity concerns.
