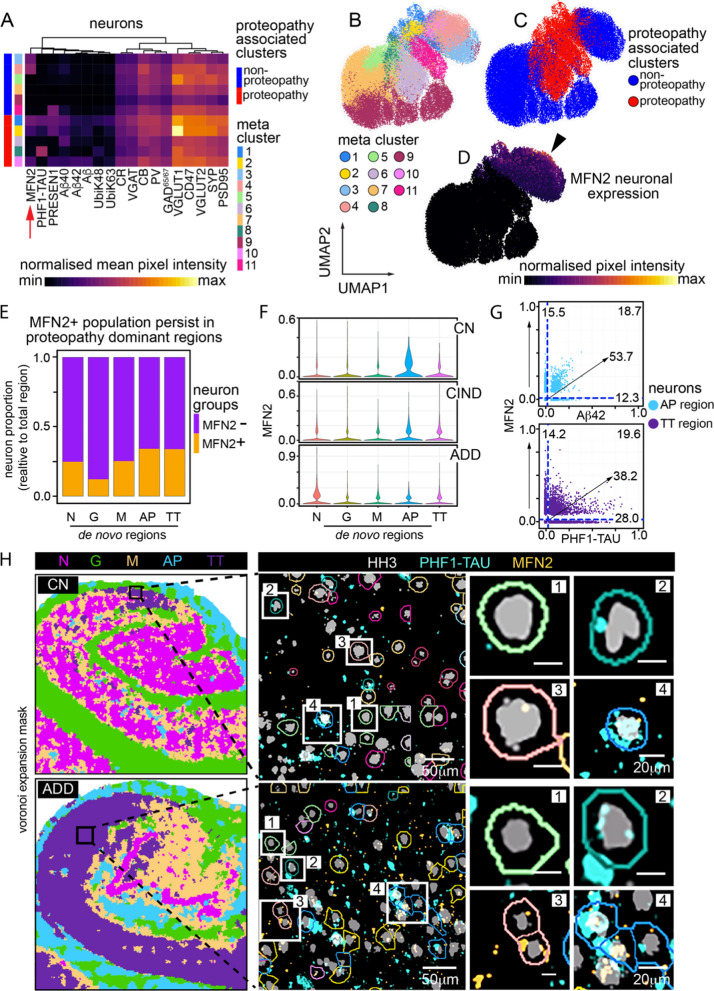
Fig. 6.
Neuronal sub-clustering reveals relationship between MFN2 and persistent neurons in AD. a Heatmap showing mean normalized expression of neuronal markers after FlowSOM clustering. 11 clusters are broadly broken down into proteopathy and non-proteopathy associated phenotypes. Red arrow denotes MFN2 cluster expression. b–d UMAP projection of neurons alone, calculated based on same markers used to perform FlowSOM clusters in a. b Overlay of neuronal subclusters. c Overlay of clusters associated with proteopathy or no-proteopathy. d Overlay of normalized neuron expression of MFN2. e Proportion of neurons positive for MFN2 vs neurons negative for MFN2 for each de novo region. f Proportions of neurons positive for MFN2 across both de novo region and AD sample condition. g Expression profile of individual neurons for MFN2 and Aβ42 in AP region (top) and MFN2 and PHF1-TAU in TT region (bottom). h Neurons in TT region of CA1, CN individual (top) and CA1, ADD individual (bottom). Black box represents areas of TT-associated tissue isolated for insets (left), zoomed in section of multiple neurons, colored by cluster ID’s in A (middle), single or small groups of neurons representing persistent, TT cells (right). [1] MFN2(−), PHF1-TAU(−) neuron—Cluster 5. [2] MFN2(−), PHF1-TAU(+) neurons—Cluster 8. [3] MFN2(+), PHF1-TAU(−) neurons—Cluster 4. [4] MFN2(+), PHF1-TAU(+) neurons—Cluster 1. Abbreviations: CN, cognitively normal; ADD, AD dementia; N, Neuronal dominant; G, Glial dominant; M, Mixed disease; AP, Aβ plaque dominant; TT, PHF1-TAU NFT-NTs dominant