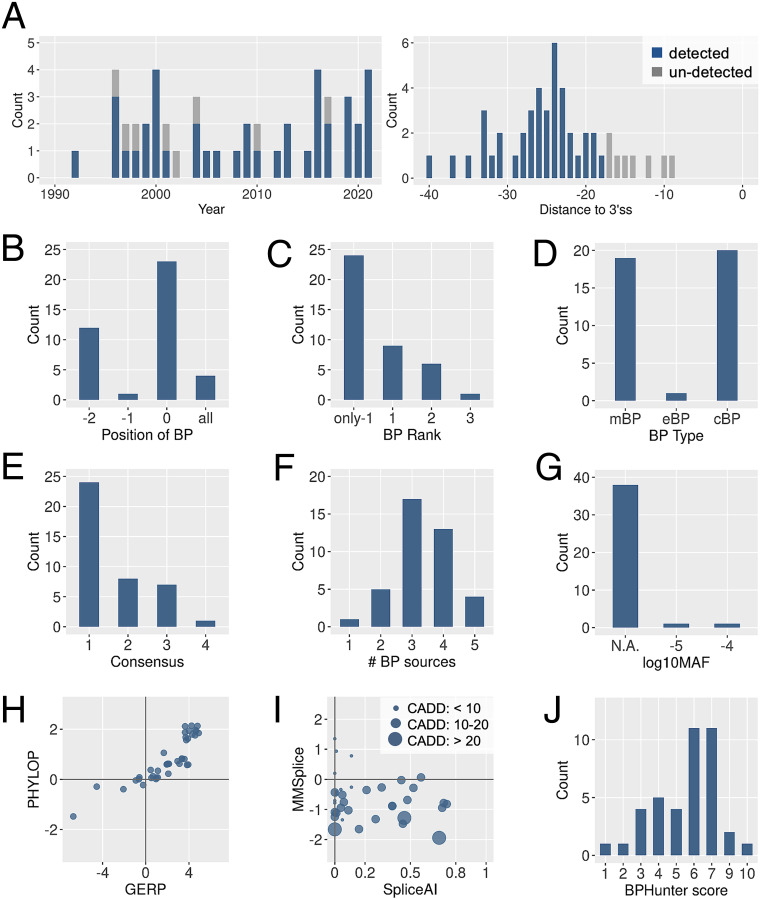
Fig. 4.
Detection of pathogenic BP variants by BPHunter. (A) Timeline of the 48 published pathogenic BP variants (Left) and the distance to their 3′ss (Right). (B) Disrupted positions of BP. (C) Ranking of the disrupted BP in its 3′-proximal intron. (D) Type of disrupted BP. (E) Level of consensus of the disrupted BP motif (1: YTNAY, 2: YTNA, 3: TNA, and 4: YNA). (F) Number of data sources supporting the disrupted BP. (G) Population variation of the disrupted BP. (H) Conservation scores of the disrupted BP. (I) Missplicing scores and deleteriousness score of the pathogenic BP variants. (J) BPHunter scores of the pathogenic BP variants.