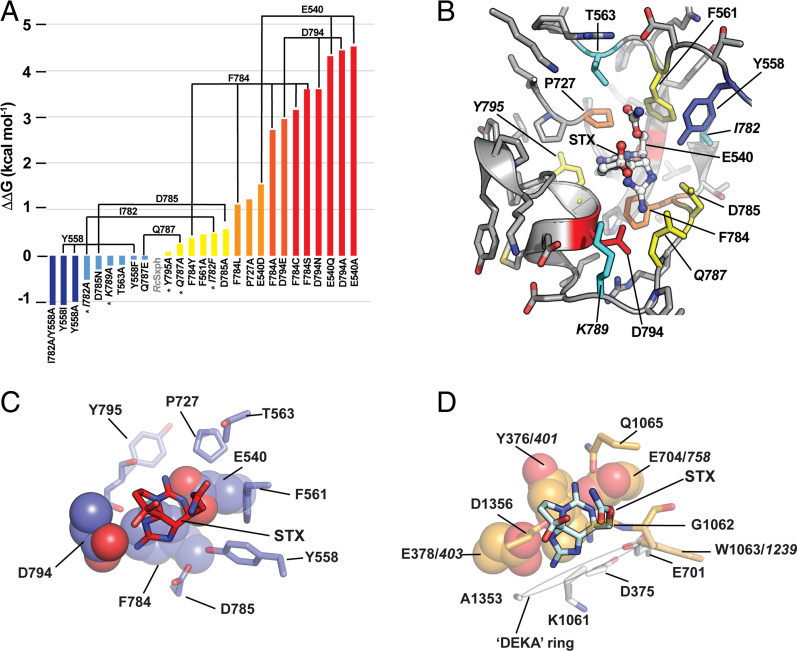
Fig. 2.
Energetic fingerprint of STX recognition by RcSxph. (A) ΔΔG comparisons for the indicated RcSxph STX binding pocket mutants relative to wild-type RcSxph. Colors indicate ΔΔG < −1 kcal⋅mol−1 (blue), −1 ≤ ΔΔG ≤ 0 kcal⋅mol−1 (light blue), 0 ≥ ΔΔG ≥ 1 kcal⋅mol−1 (yellow), 1 ≥ ΔΔG ≥ 2 kcal⋅mol−1 (orange), 2 ≥ ΔΔG ≥ 3 kcal⋅mol−1 (red orange), and ΔΔG ≥ 3 (red). (B) Energetic map of alanine scan mutations on STX binding to the RcSxph STX binding pocket (PDB ID:6O0F) (10). Second-shell sites are in italics. Colors are as in A. (C and D) Structural interactions of STX with (C) RcSxph (PDB ID:6O0F) (10) and (D) human NaV1.7 (PDB ID:6J8G) (40). Residues that are energetically important for the STX interaction are shown in space filling. NaV1.7 selectivity filter “DEKA” ring residues are shown (white). Italics indicate corresponding residue numbers for rat NaV1.4 (18).