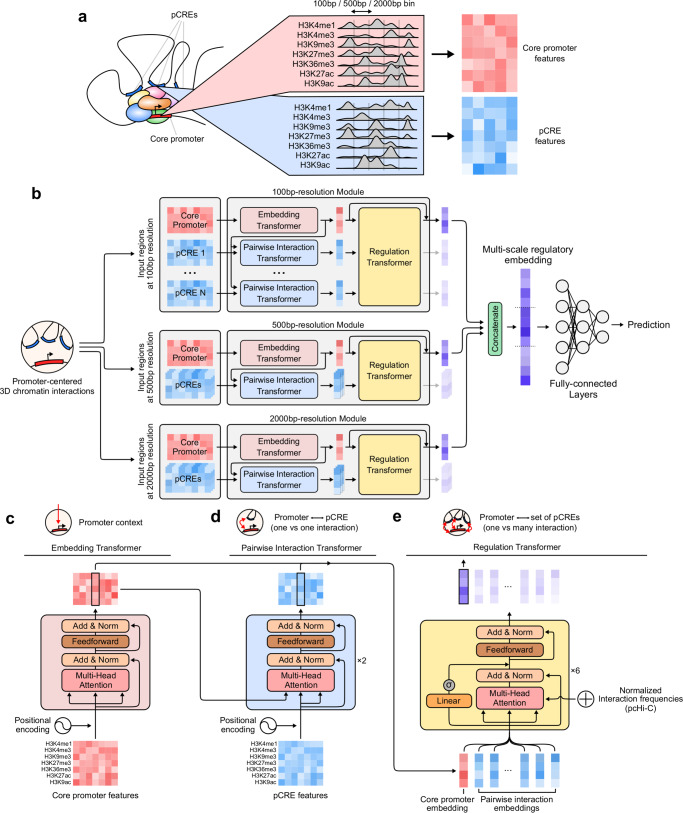
Fig. 1. Chromoformer model architecture.
a Input features. To predict the expression of a gene using levels of histone modifications (HMs), we extracted binned average signals of HMs from both the core promoter and putative cis-regulatory regions (pCREs). b Chromoformer architecture. Three independent modules were used to produce multi-scale representation of gene expression regulation. Each of the modules is fed with input HM features at different resolutions to produce an embedding vector reflecting the regulatory state of the core promoter. c Embedding transformer architecture. Position-encoded HM signals of core promoter features are transformed into core promoter embeddings through self-attention. d Pairwise Interaction transformer architecture. Position-encoded HM signals of pCREs are used to transform the core promoter embeddings into Pairwise Interaction embeddings through encoder-decoder attention. e Regulation transformer architecture. Using the whole set of the core promoter and Pairwise Interaction embeddings and gated self-attention, the Regulation transformer learns how the pCREs collectively regulate the core promoter. To guide the model to put greater attention to frequently occurring three-dimensional (3D) interactions, the normalized interaction frequency vector is added to self-attention affinity matrices.