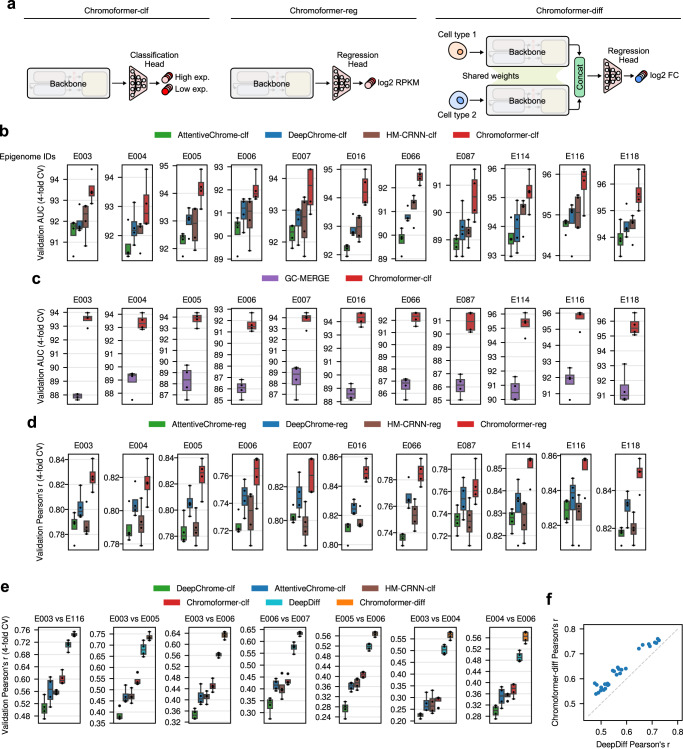
Fig. 2. Chromoformer outperformed existing deep learning models in predicting gene expression in various aspects.
a Three variants of Chromoformer evaluated in this study. Chromoformer-classifier (Chromoformer-clf) predicts binary gene expression labels. Chromoformer-regressor (Chromoformer-reg) predicts the expression levels (in log2 RPKM) of genes. Chromoformer-diff predicts the log2 fold-change of gene expression given the HM configurations of two different cell types for each gene. b Cross-validation (n = 4) performances of Chromoformer-clf compared to the benchmark deep learning models that only utilize the core promoter features. c Comparison of cross-validation (n = 4) performances with GC-MERGE, a graph neural network model that utilizes 3D cis-regulatory interactions. For fair comparisons, Chromoformer-clf models were retrained from scratch using only a subset of genes that GC-MERGE can predict. d Cross-validation (n = 4) performances of Chromoformer-reg compared to the benchmark deep learning models. Prediction heads of benchmark models were modified to produce single scalar values instead of binary labels. e Cross-validation (n=4) performances of Chromoformer-diff compared to the benchmark deep learning models. Evaluation of the fold-change prediction based on the ratio of classification probabilities are shown as a reference (denoted as DeepChrome-clf, AttentiveChrome-clf, HM-CRNN-clf, and Chromoformer-clf). f Pairwise performance comparison between DeepDiff and Chromoformer-diff. Throughout b–e the center line denotes the median, the upper and lower box limits denote upper and lower quartiles, and the whiskers denote 1.5× interquartile range. AUC, Area under the receiver operating characteristic curve; CV, Cross-validation. Source data are provided as a Source Data file.