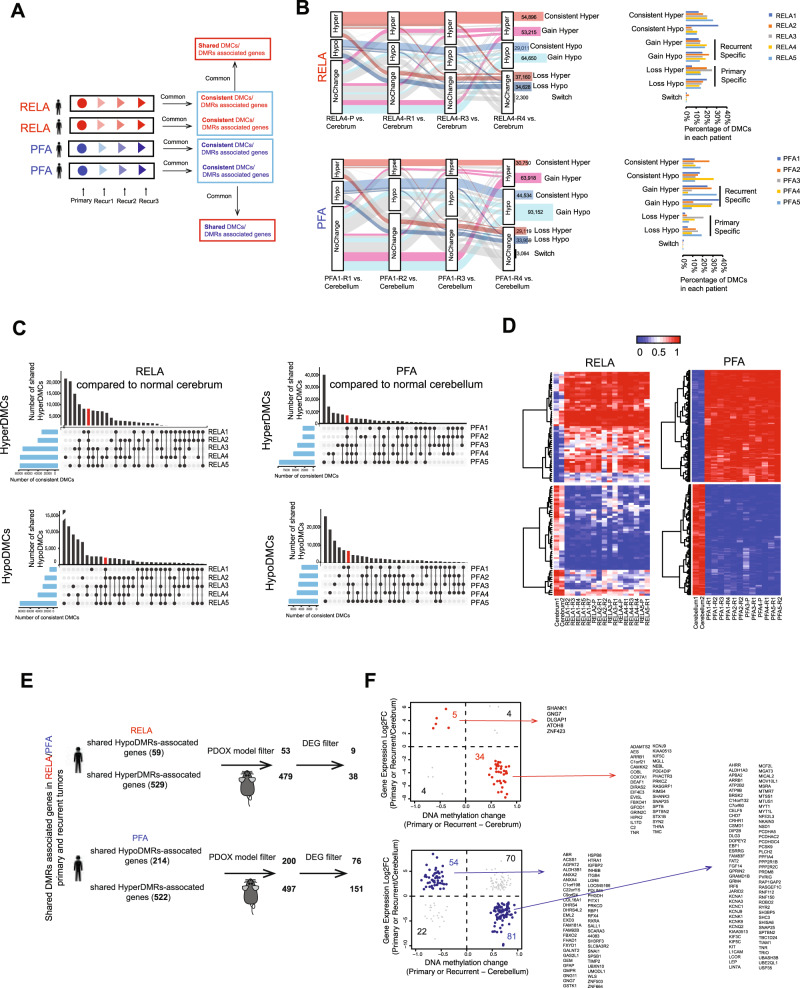
Fig. 3. Identification of potential DNA methylation drivers for ependymoma relapse.
A Schematic illustration of dynamic analysis of time-course DNA methylation to identify consistent DMCs that were present from primary to all recurrent tumors in each individual patient, and the shared DMCs that were consistently present in all RELA or PFA patients. B Representative alluvia plots (left panel) showing the dynamic changes of DNA methylation for RELA4 (left upper panel) and PFA1 (left lower panel) tumors during repeated recurrences. Normal brain tissues were used as reference to determine the CpG status, i.e., Hyper-, Hypo-methylation and No Change. Seven different patterns (categories) and the numbers of CpG changes were listed with different colors. Graphs (right panel) showed percentages of the 7 different categories of CpG changes of each RELA (right upper panel) and PFA patient (right lower panel). C UpSet R plots showing the number of consistent Hyper- and HypoDMCs (y-axis) that were shared among RELA (left panels) and PFA patients (right panels) (connected dots with lines) as well as numbers of consistent DMCs for each patient (the horizonal histograms). Consistent DMCs that were shared by all RELA or PFA accounted for a small fraction and were highlighted in red, respectively. D Heatmaps showing the DNA hyper- (red) and hypo-methylation (blue) ratios of potential DNA methylation drivers (CpGs) for RELA (left panels) and PFA (right panels) recurrent tumors. E Schematic illustration of the data analysis steps to identify potential driver genes regulated by potential DNA methylation drivers. Differentially expressed genes (DEGs) (log2 fold change between tumor and normal tissues) were extracted from RNA-seq of the same set of tumors. DEGs discovered in patient tumors but absent in the matching PDOX tumors were filtered out to identity genes that contributed to PDOX tumorigenicity. F Correlation of expression and DNA methylation of potential genes were shown in the scatterplots for RELA (upper panel) and PFA (the lower panel). Those negatively correlated in RELA were highlighted in red (upper panel) and in PFA in blue (lower panel) and listed to the right of the plot.