Abstract
Aims
Cardiotoxicity leading to heart failure (HF) is a growing problem in many cancer survivors. As specific treatment strategies are not available, RNA discovery pipelines were employed and a new and powerful circular RNA (circRNA)-based therapy was developed for the treatment of doxorubicin-induced HF.
Methods and results
The circRNA sequencing was applied and the highly species-conserved circRNA insulin receptor (Circ-INSR) was identified, which participates in HF processes, including those provoked by cardiotoxic anti-cancer treatments. Chemotherapy-provoked cardiotoxicity leads to the down-regulation of Circ-INSR in rodents and patients, which mechanistically contributes to cardiomyocyte cell death, cardiac dysfunction, and mitochondrial damage. In contrast, Circ-INSR overexpression prevented doxorubicin-mediated cardiotoxicity in both rodent and human cardiomyocytes in vitro and in a mouse model of chronic doxorubicin cardiotoxicity. Breast cancer type 1 susceptibility protein (Brca1) was identified as a regulator of Circ-INSR expression. Detailed transcriptomic and proteomic analyses revealed that Circ-INSR regulates apoptotic and metabolic pathways in cardiomyocytes. Circ-INSR physically interacts with the single-stranded DNA-binding protein (SSBP1) mediating its cardioprotective effects under doxorubicin stress. Importantly, in vitro transcribed and circularized Circ-INSR mimics also protected against doxorubicin-induced cardiotoxicity.
Conclusion
Circ-INSR is a highly conserved non-coding RNA which is down-regulated during cardiotoxicity and cardiac remodelling. Adeno-associated virus and circRNA mimics-based Circ-INSR overexpression prevent and reverse doxorubicin-mediated cardiomyocyte death and improve cardiac function. The results of this study highlight a novel and translationally important Circ-INSR-based therapeutic approach for doxorubicin-induced cardiac dysfunction.
Keywords: Heart failure • Circular RNA • Doxorubicin cardiotoxicity • AAVtherapy • Mitochondrial metabolism • Anti-cancer treatment
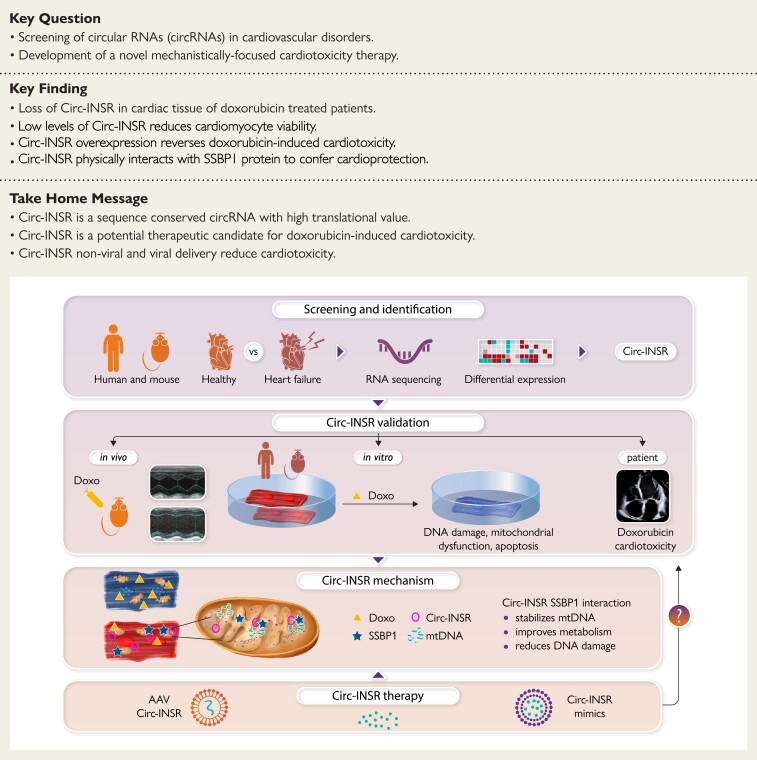
Structured Graphical Abstract
Structured Graphical Abstract.
Circular RNA therapy for cardioprotection.
AAV, adeno-associated virus; INSR, insulin receptor; SSBP1, single-stranded DNA-binding protein.
See the editorial comment for this article ‘‘Circulating' RNA-based therapies in Cardio-Oncology’, by Carlo G. Tocchetti et al., https://doi.org/10.1093/eurheartj/ehac407.
Translational perspective.
The recent advances in anti-cancer treatments have led to a steady increase in the number of cancer survivors, paradoxically also enhancing the number of heart failure (HF) patients who suffer from the undesired cardiotoxic side effects of these anti-tumour treatments. Therapeutic options to reduce or reverse chemotherapy-induced HF are unavailable. Circular RNAs exhibit high druggability due to their high stability arising from special closed loop structure. Circ-INSR overexpression prevents doxorubicin-induced cardiac dysfunction in pre-clinical murine and human models. Furthermore, Circ-INSR mimics reduced cardiomyocyte death under doxorubicin stress, highlighting the potential of Circ-INSR-based RNA therapy for future clinical applications.
Introduction
Cardiovascular disease (CVD) as the leading cause of death results in an immense health and economic burden globally.1 Cardiomyocyte death and adverse cardiac remodelling, which ultimately progress into heart failure (HF), are a frequent outcome induced by anti-cancer drug treatments leading to the death of the patient.2–4 Due to novel and improved anti-cancer treatments, the number of cancer survivors is steadily increasing5,6 which, paradoxically, increases the number of HF patients as a result of the undesired side effects of these treatments.7 A perfect example is doxorubicin, an anthracycline class drug, which was identified as a very powerful anti-cancer medication that has been applied in clinics since the late 1960s.8 However, both acute and long-term adverse effects, especially on the heart leading to myocardial damage and HF, limit doxorubicin administration to cancer patients who may otherwise strongly benefit from this treatment and other chemotherapeutic agents of its kind.9,10 This complex dilemma is addressed by the new field of cardio-oncology, which explores novel therapeutic strategies for the treatment of anti-cancer drug-induced HF and, most importantly, novel therapeutic targets for the primary and secondary prevention of cardiotoxic effects, to avoid the development of HF. In this regard, non-coding RNAs (ncRNAs), particularly microRNAs and long ncRNAs, have emerged as powerful targets for the prevention and treatment of CVD in pre-clinical models and beyond.11–15 Indeed, the first human clinical trial using ncRNA modulator in HF patients showed encouraging results, thus highlighting the translational potential for the next generation of RNA-based HF therapeutics.13,16 In addition to microRNAs and long ncRNAs, a recently discovered class of ncRNA molecules, circular RNAs (circRNAs), are beginning to gain more focus as novel therapeutic targets for CVDs.17 Characterized by covalently linked ends, thus forming closed RNA circles, these circRNAs possess higher levels of stability when compared with linear RNA.18,19 The CircRNAs are mainly formed through non-canonical splicing of pre-mRNA transcripts, a process known as back splicing.20 They are abundant in certain tissue types and diseases and often seen to be highly conserved across species, which facilitates their pre-clinical investigation and underlines their translational potential.21,22 There is experimental evidence describing that circRNAs are indeed functional molecules with several modes of action.17,18 These include the ability to act as a molecular sponge for microRNAs or RNA-binding proteins, to provide a scaffold function for protein complexes, to regulate cellular processes such as nuclear-cytoplasmic shuttling, and to encode for functional micro-peptides.23–25 However, the mechanisms of circRNAs in HF or cardiotoxicity are still poorly understood and await in-depth studies.
Here, we investigated novel circRNAs as potential therapeutic targets for HF specifically in the context of doxorubicin-induced cardiac dysfunction. Our circRNA sequencing approach identified a novel, highly abundant, and sequence conserved circRNA molecule derived from the host gene encoding the insulin receptor (INSR). While INSR signalling is known to play an important role in cardiac homeostasis and disease,26 we found that the circRNA derived from this locus, Circ-INSR, is regulated by breast cancer Type 1 susceptibility protein (Brca1) independently of the host gene expression. We applied various gain- and loss-of-function studies specific for Circ-INSR to validate its cardioprotective effects and elucidate the underlying mode of action both in vitro and in vivo. We also succeeded in producing of in vitro transcribed Circ-INSR mimics, which can be used as a fast and efficient method to deliver the circRNA molecule to initiate the therapeutic process. In this study, we discovered Circ-INSR as a promising target for the prevention of anti-cancer drug-induced cardiotoxicity.
Methods
Detailed methods are provided in the Supplementary material online.
Human tissues sampling
Cardiac tissue samples included those from patients with HF,27 doxorubicin-induced HF, or healthy controls.12 This study (using failing hearts and healthy controls) was performed with the approval of the institutional ethics committee of the Hannover Medical School, Germany. Collection of the human heart tissue (doxorubicin anti-cancer therapy) was approved by the scientific advisory board of the biobank of the University Medical Center Utrecht, Utrecht, The Netherlands and the University Medical Center Groningen, Groningen, The Netherlands (Protocol Nos 12/387 and 15/252). The study was performed in accordance with the guidelines from the declaration of Helsinki and its amendments or comparable ethical standards. For details, see Supplementary material online.
Animal experiment
All animal experiments were approved by the local authorities at Hannover Medical School and Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, LAVES. For details, see Supplementary material online.
Cell culture
Cardiomyocyte-like, HL-1 cells, HEK-293T cells, neonatal mouse/rat cardiomyocytes, and human-induced pluripotent stem-cell (hiPSC) line Phoenix (hHSC_Iso4_ADCF_SeV-iPS2, alternative name: MHHi001-A) were cultured according to standard protocols. Phoenix hiPSCs were differentiated into cardiomyocytes using Wnt pathway modulators using a previously established cardiomyocyte differentiation protocol.28,29 For details, see Supplementary material online.
Circular RNA overexpression plasmid strategy
Murine Circ-INSR sequence and the circularization elements were polymerase chain reaction (PCR) amplified from HL-1 cell genomic DNA using Hot Star Taq Master Mix (Qiagen). EcoRI and Xhol sites present in the forward and reverse primers were used to add restriction enzyme sites to the amplified transgene, which was subcloned into the pcDNA3.1(+) Laccase2 MCS Exon Vector (Addgene #69893) cloning. For details, see Supplementary material online.
In vitro RNA transcription and circularization
In vitro RNAs were produced and modified by using RiboMax large RNA production system (Promega) according to the manufacturer’s protocol. Briefly, 5 μg PCR-amplified T7 DNA fragments were incubated in 100 μL reaction system with 10 μL T7 RNA polymerase enzyme mix, 25 mM ATP, 25 mM CTP, 25 mM GTP, 20 mM GMP, and 5 mM GTP at 37°C for 2 h, followed by DNase I treatment for 15 min at 37°C to remove DNA templates. The GMP is required for the production of 5′-monophosphate RNA and subsequent RNA circularization. In vitro RNAs were precipitated with 3 M NaOAc (pH 5.2) and isopropyl alcohol, washed with 70% ethanol and resuspended in RNase-free water. One micromolar linear RNAs were incubated with 3 μM DNA splint oligomer (complementary 10-nt regions at the 5′ and 3′ ends of the RNA), T4 DNA ligase (Thermo Fisher) in 500 μL reaction for 2 h according to the published protocol.30
Statistics
All data were analysed using GraphPad Prism software. Data are presented as mean ± standard error mean (SEM), and an unpaired two-tailed t-test or Mann–Whitney U test was performed to calculate significance between two groups, one-way analysis of variance (ANOVA) with post hoc Tukey test or Bonferroni’s multiple comparison test was used to calculate significance between ≥3 groups, and two-way ANOVA with Sidak’s or Tukey’s multiple comparison test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant.
Results
Identification and validation of the conserved circular RNA, Circ-INSR, in heart failure
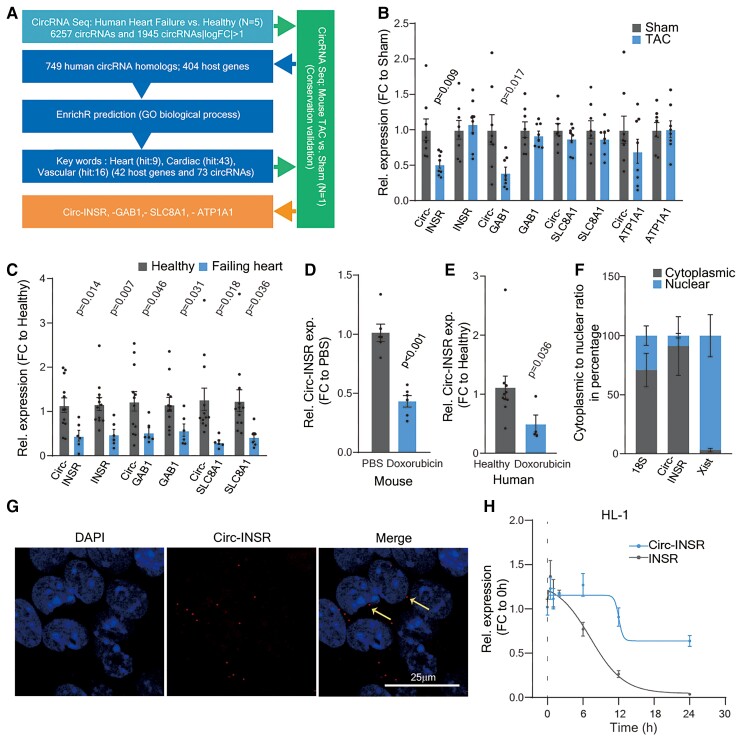
We performed global circRNA profiling in left ventricular cardiac tissue from healthy humans and HF patients and from mouse hearts with and without left ventricular pressure overload-induced cardiac remodelling. A total of 6257 circRNAs were detected in human hearts by circRNA sequencing, of which 1945 circRNAs were differentially expressed (fold change >2) between healthy and failing human left ventricles (Supplementary material online, Figure S1A). We focused on highly conserved circRNAs between mice and humans, and detected 749 conserved circRNA homologs, which were derived from 404 host genes. As circRNAs are back spliced from precursor mRNAs, we hypothesized that circRNAs could have comparable function to their host mRNAs.
We thus applied gene ontology (GO) analyses to pinpoint circRNAs potentially involved in cardiovascular processes. We identified four highly conserved circRNAs whose host genes are crucially involved in cardiac function, namely Circ-INSR (mmu-circ-0014773 and hsa-circ-0000885), Circ-ATP1A1 (mmu_circ_0010303 and hsa_circ_0013692), Circ-SLC8A1 (mmu_circ_0000823 and hsa_circ_0120059), and Circ-GAB1 (mmu_circ_0015017 and hsa_circ_0125422; Figure 1A). We validated the expression of these circRNAs in murine failing hearts and human hearts from HF patients (Figure 1B and C). As Circ-INSR displayed a significant decrease in both rodent and human failing heart tissue, we selected this candidate to further explore its function in other HF models. The Circ-INSR was additionally down-regulated in a doxorubicin-mediated cardiotoxicity mouse model and in cardiac tissue from patients who developed cardiotoxicity after anti-cancer treatment with doxorubicin (Figure 1D and E). This repression could be consistently recapitulated in vitro after treatment of murine HL-1 cardiomyocyte-like cells and human-induced pluripotent stem-cell-derived cardiomyocytes with doxorubicin (Supplementary material online, Figure S1B and C).
Figure 1.
Identification of Circ-INSR from failing hearts and validation in various heart failure models. (A) Pipeline to select candidates from RNA-Seq data. (B) Relative expression of circRNA and their linear host genes (Circ-INSR, INSR, Circ-GAB1, GAB1, Circ-SLC8A1, SLC8A1, Circ-ATP1A1, and ATP1A1) in TAC model at 13 weeks (n = 8 mice/group). (C) Relative expression of circRNA and their linear host genes (Circ-INSR, INSR, Circ-GAB1, GAB1, Circ-SLC8A1, and SLC8A1) in failing heart (n = 6 patients) compared with healthy (n = 12), expression of Circ-ATP1A1 was not detectable in the human samples. (D) Relative expression of Circ-INSR in mouse hearts treated with doxorubicin (n = 6 per group). (E) Relative expression of Circ-INSR in male patient heart with doxorubicin-induced heart failure (n = 4) compared with healthy (n = 10; Mann–Whitney U test was performed). (F) Circ-INSR expression in neonatal rat cardiomyocytes represented after normalization to the respective subcellular fraction (18S applied for normalization of cytoplasmic fraction, Xist applied for normalization of nuclear fraction, n = 6 per group from duplicates of three independent experiments). (G) RNA fluorescence in situ hybridization (RNA-FISH) of Circ-INSR in HL-1 cells (scale bar, 25 μm). Thin yellow arrows indicate cytoplasmic localization of the Circ-INSR as observed in RNA-FISH experiment. (H) Relative expression of Circ-INSR and INSR in HL-1 cells treated with Actinomycin D (n = 6 per timepoint). All quantitative data are presented as mean ± standard error mean, and an unpaired two-tailed t-test or Mann–Whitney U test was performed to calculate significance between two groups. P < 0.05 was considered as statistically significant. exp., expression; FC, fold change; GO, gene ontology; NRCM, neonatal rat cardiomyocyte; TAC, transverse aortic constriction; Rel., relative.
We validated the high sequence conservation of Circ-INSR using NCBI BLAST analysis (human vs. mouse 85.87%, human vs. rat 86.41%, rat vs. mouse 95.83%; Supplementary material online, Figure S1D). Next, the back-splicing sites were confirmed by Sanger sequencing of PCR products generated with Circ-INSR-specific divergent primers in different species (Supplementary material online, Figure S1E). Increased resistance against exonucleolytic digest by RNaseR and the lack of polyA sequences, two important characteristics of circRNAs,17 further confirmed the circular structure of Circ-INSR (Supplementary material online, Figure S1F and G). Subcellular fractionation and RNA-FISH experiments revealed a predominant cytoplasmic localization of Circ-INSR within the cell (Figure 1F and G). Treatment of HL-1 and neonatal rat cardiomyocytes with the transcription blocker actinomycin D revealed a longer half-life of Circ-INSR compared with linear INSR (Figure 1H and Supplementary material online, Figure S1H). Circ-INSR is highly enriched in human-induced pluripotent stem-cell-derived cardiomyocytes compared with human umbilical vein endothelial cells and human cardiac fibroblasts (Supplementary material online, Figure S1I). Collectively, these data validate Circ-INSR to be sensitive to doxorubicin treatment and thus it may function as a potential therapeutic target in doxorubicin-induced HF.
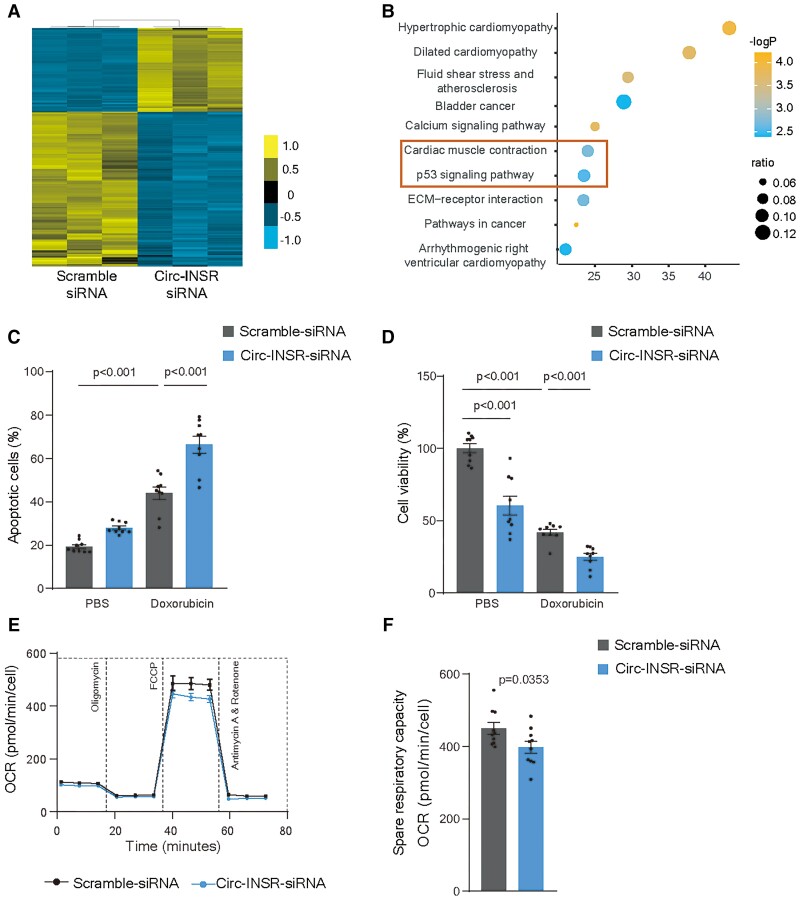
Lack of Circ-INSR impairs metabolic activity and survival of cardiomyocytes
To gain further functional insights, we designed siRNAs which specifically targeted the back-splicing sequence of Circ-INSR degrading the circular transcript without affecting the linear INSR transcript. We found two siRNAs that explicitly degraded Circ-INSR but not the linear transcript (Supplementary material online, Figure S2A and B). Interestingly, INSR was up-regulated after Circ-INSR siRNA2 transfection in HL-1 cells. Therefore, we selected siRNA1 for targeting mouse Circ-INSR in the following experiments. We performed RNA sequencing in HL-1 cardiomyocytes with and without Circ-INSR silencing. We identified 1376 genes to be differentially expressed (P < 0.05, fold change >2) after Circ-INSR knockdown (Figure 2A). The KEGG analysis pointed to the regulation of genes involved in cardiac biology and metabolic processes (Figure 2B). We next tested the functional involvement of Circ-INSR in doxorubicin-mediated cardiotoxicity and found silencing of Circ-INSR exaggerated the doxorubicin-induced cardiotoxicity (Figure 2C and Supplementary material online, Figure S2C). In line, cellular viability was reduced under doxorubicin stress and deteriorated further after Circ-INSR knockdown (Figure 2D). As RNA sequencing suggested metabolic pathways to be regulated by Circ-INSR, we analysed the metabolic activity of neonatal mouse cardiomyocytes after siRNA-mediated Circ-INSR down-regulation. Circ-INSR silencing (Supplementary material online, Figure S2D) negatively influenced mitochondrial respiration as indicated by lower oxygen consumption rate and reduced spare respiratory capacity (Figure 2E and F). Collectively, these data demonstrate that loss of Circ-INSR in cardiomyocytes leads to impaired metabolic activity and induces apoptosis during doxorubicin stress, suggesting that Circ-INSR possesses cardioprotective effects.
Figure 2.
Down-regulation of Circ-INSR reduces cardiomyocytes survival and disrupts metabolic activity. (A) Heat map of RNA-Seq data for selected genes from the HL-1 cells treated with Circ-INSR-siRNA compared with Scramble siRNA (blue indicates down-regulated genes, yellow indicates up-regulated genes). (B) KEGG analysis of the dysregulated gene pathways from the RNA sequencing data. Orange box in the bubble plot highlights the most prominently regulated cellular processes after Circ-INSR knockdown. (C) Annexin-V and 7-AAD staining of Circ-INSR-siRNA transfected HL-1 cells with/without doxorubicin treatment (0.25 μM 48 h, n = 9 wells per group from triplicates of three independent experiments, one-way analysis of variance was performed within Scramble siRNA, Scramble siRNA + Doxorubicin, and Circ-INSR-siRNA + Doxorubicin). (D) Percentage of viable cells (WST-1 assay) in Circ-INSR-siRNA treated HL-1 cells compared with Scramble siRNA with/without doxorubicin treatment (0.25 μM 48 h, n = 9 wells per group from triplicates of three independent experiments, one-way analysis of variance was performed within Scramble siRNA, Scramble siRNA + Doxorubicin, and Circ-INSR-siRNA + Doxorubicin, unpaired two-tailed t-test was performed between Scramble siRNA and Circ-INSR-siRNA). (E) Mean oxygen consumption rate at various time points of Mito stress assay (n = 10 wells per group from one experiment). (F) Spare respiratory capacity of neonatal mouse cardiomyocytes treated with Circ-INSR-siRNA compared with Scramble siRNA (n = 10 wells per group from one experiment). All quantitative data are presented as mean ± standard error mean, and an unpaired two-tailed t-test was performed to calculate significance between two groups, and one-way analysis of variance with post hoc Tukey test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant. NMCM, neonatal mouse cardiomyocyte; OCR, oxygen consumption rate.
Circ-INSR therapy prevents doxorubicin-induced metabolic defects and apoptosis in cardiomyocytes
To test whether counteracting endogenous down-regulation of Circ-INSR could reverse doxorubicin-induced cardiotoxicity, we constructed a Circ-INSR overexpression cassette consisting of the Circ-INSR exon flanked by intronic sequences containing circularization elements (Supplementary material online, Figure S3A). The overexpression construct was subcloned into the pcDNA backbone followed by transfection into HL-1 cardiomyocytes after which the successful up-regulation of Circ-INSR was confirmed by RT-qPCR and agarose gel electrophoresis (Supplementary material online, Figure S3B). To induce Circ-INSR expression in primary cardiomyocytes, the overexpression cassette was subcloned into the adeno-associated virus (AAV) backbone and packaged into AAV6 viral particles allowing efficient transduction of Circ-INSR in primary rat and mouse cardiomyocytes (Supplementary material online, Figure S3C and D). Overexpression of Circ-INSR in HL-1 cardiomyocytes attenuated doxorubicin-mediated apoptosis and enhanced cell viability (Figure 3A and B, Supplementary material online, Figure S3E). In line, overexpression of Circ-INSR in primary rat cardiomyocytes also attenuated doxorubicin-induced DNA damage and apoptosis (Figure 3C and D). Notably, overexpression of Circ-INSR increased mitochondrial performance indicated by improved oxygen consumption rate, basal and spare respiratory capacity as well as maximal respiration both under basal conditions or after doxorubicin treatment (Figure 3E–G, Supplementary material online, Figure S3F).
Figure 3.
Circ-INSR overexpression alleviates doxorubicin-induced metabolic disorder and cardiomyocyte apoptosis. (A) Percentage of apoptotic cells quantified by Annexin-V and 7-AAD staining of pcDNA-Empty and pcDNA-Circ-INSR transfected HL-1 cells with/without doxorubicin (0.25 μM 48 h, n = 12/6/19/18 from three independent experiments, one-way analysis of variance was performed within pcDNA-Empty, pcDNA-Empty + Doxorubicin, and pcDNA-Circ-INSR + Doxorubicin). (B) Percentage of viable HL-1 cells (WST-1 assay) in pcDNA-Circ-INSR overexpression HL-1 cells compared with pcDNA-Empty control (0.25 μM 48 h, n = 12/8/8/10 from three independent experiments, one-way analysis of variance was performed within pcDNA-Empty, pcDNA-Empty + Doxorubicin, and pcDNA-Circ-INSR + Doxorubicin, unpaired two-tailed t-test was performed between pcDNA-Empty and pcDNA-Circ-INSR). (C) γ-H2AX (red) staining in nuclei (DAPI, blue) of neonatal rat cardiomyocytes (cTNT, green) transduced with either AAV6-Empty control or AAV6-Circ-INSR, in the presence or absence of doxorubicin. Yellow arrows denote DNA damage (0.25 μM 48 h, n = 8/6/8/8 from three independent experiments, one-way analysis of variance was performed within AAV6-Empty, AAV6-Empty + Doxorubicin, and AAV6-Circ-INSR + Doxorubicin). Representative images left; quantification right (scale bar, 50 μm). (D) TUNEL positive (green) staining in nuclei (DAPI, blue) of neonatal rat cardiomyocytes (cTNT, red) transduced with either AAV6-Empty control or AAV6-Circ-INSR, in the presence or absence of doxorubicin. Yellow arrows indicate apoptotic cells (0.25 μM 48 h, n = 6/5/6/6 from three independent experiments, one-way analysis of variance was performed within AAV6-Empty, AAV6-Empty + Doxorubicin, and AAV6-Circ-INSR + Doxorubicin); Representative images left; quantification right (scale bar, 50 μm). (E) Mean oxygen consumption rate at various time points of Mito Stress Assay (5 μM, 2 h, n = 17/16/16/16 wells from one experiment). (F and G) Basal respiration and spare respiratory capacity of NMCMs treated with AAV6-Circ-INSR compared with AAV6-Empty under basal conditions or doxorubicin treatment (5 μM, 2 h, n = 17/16/16/16 wells from one experiment; one-way analysis of variance (Bonferroni’s multiple comparison test) was performed within AAV6-Empty, AAV6-Empty + Doxorubicin and AAV6-Circ-INSR + Doxorubicin, unpaired two-tailed t-test was performed between AAV6-Empty and AAV6-Circ-INSR). All quantitative data are presented as mean ± standard error mean, and one-way analysis of variance with post hoc Tukey test or Bonferroni’s multiple comparison test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant. FC, fold change; NRCM/NMCM, neonatal rat/mouse cardiomyocyte; OCR, oxygen consumption rate.
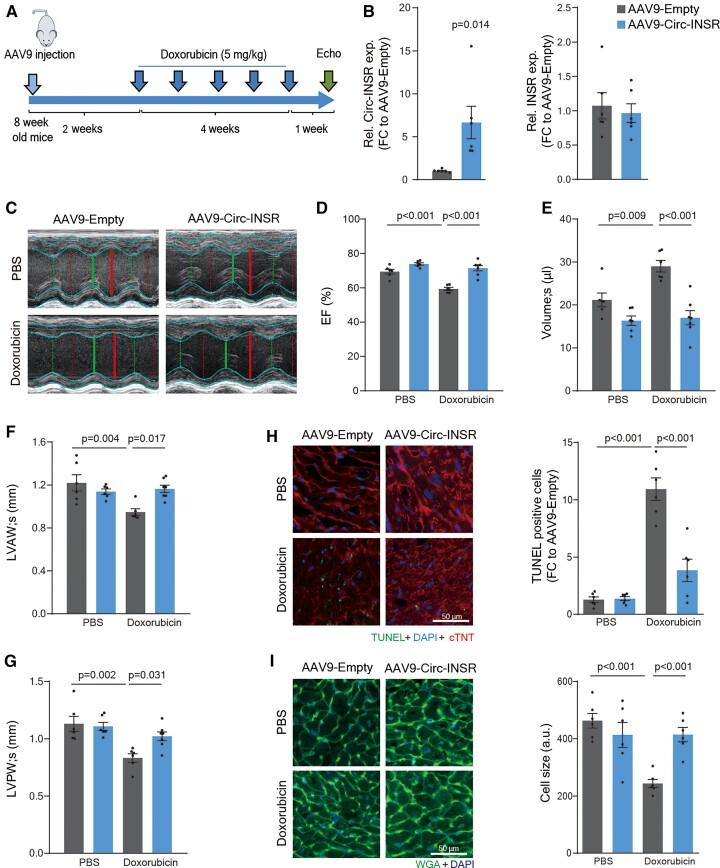
Design and efficacy of Circ-INSR therapy in a murine in vivo model of doxorubicin-induced cardiotoxicity
To test the therapeutic potential of Circ-INSR in vivo, we employed a previously established mouse model of chronic doxorubicin-induced cardiotoxicity.29,31,32 We produced AAV9-Circ-INSR and control AAV9-Empty viral particles and injected them into adult male mice (8 weeks) followed by chronic doxorubicin treatment for 5 weeks (cumulative dose 25 mg/kg) (Figure 4A). In this model, we confirmed the previously identified doxorubicin-induced down-regulation of Circ-INSR (Figure 1D and E), while AAV9-Circ-INSR-mediated treatment led to a six-fold cardiac up-regulation of Circ-INSR expression (Figure 4B). No change in cardiac expression of the linear INSR mRNA was observed assuring specific overexpression of the Circ-INSR only (Figure 4B). Cardiac function analyses by echocardiography showed a significant reduction of left ventricular ejection fraction in AAV9-Empty control-treated mice chronically treated with doxorubicin. Notably, cardiac function was prevented in mice treated with AAV9-Circ-INSR (Figure 4C and D). Chronic doxorubicin treatment induced adverse cardiac remodelling such as increase in left ventricular end-systolic volume as well as cardiac atrophy measured by ventricular wall thinning, which was significantly improved in mice treated with AAV9-Circ-INSR compared with AAV9-Empty controls (Figure 4E–G). A complete overview of the echocardiography analyses is provided in Supplementary material online, Table S1. Histopathological analyses of transverse heart sections from these mice, revealed a strong induction of cardiomyocyte apoptosis in response to doxorubicin, which was blunted in mice receiving AAV9-based Circ-INSR treatment (Figure 4H). Doxorubicin-induced cardiac atrophy was completely prevented by AAV9-based Circ-INSR overexpression suggesting cardiac protection of these animals (Figure 4I).
Figure 4.
Circ-INSR therapy protects from doxorubicin-induced cardiotoxicity in vivo. (A) Schematic representation of the in vivo animal model of chronic doxorubicin-induced heart failure. (B) Relative expression of Circ-INSR and INSR in AAV9-Circ-INSR and AAV9-Empty heart tissue (n = 6 mice/group). (C) Representative figures of echocardiography (Echo) after AAV9-Circ-INSR therapy in presence or absence of doxorubicin cardiotoxicity. (D–G) Ejection fraction, left ventricular end-systolic volume (volume; s), dimension (in mm) of the anterior wall of the left ventricle at systole (s), dimension (in mm) of the posterior wall of the left ventricle at systole (s) in AAV9-Circ-INSR and AAV9-Empty-treated mice with or without doxorubicin (n = 6/6/6/7 mice, one-way analysis of variance was performed within AAV9-Empty, AAV9-Empty + Doxorubicin, and AAV9-Circ-INSR + Doxorubicin). (H) TUNEL (green) co-staining with DAPI in nuclei (blue) and cTNT (red) in AAV9-Empty and AAV9-Circ-INSR heart sections with/without doxorubicin treatment; representative images left; quantification right (n = 6 mice/group, scale bar, 50 μm, one-way analysis of variance was performed within AAV9-Empty, AAV9-Empty + Doxorubicin, and AAV9-Circ-INSR + Doxorubicin). (I) WGA (green) staining in AAV9-Empty and AAV9-Circ-INSR heart sections with/without doxorubicin treatment. Blue staining indicates DAPI-stained nuclei. Representative images left; quantification right (n = 6 mice/group, scale bar, 50 μm, one-way analysis of variance was performed within AAV9-Empty, AAV9-Empty + Doxorubicin, and AAV9-Circ-INSR + Doxorubicin). All quantitative data are presented as mean ± standard error mean, and an unpaired two-tailed t-test was performed to calculate significance between two groups, and one-way analysis of variance with post hoc Tukey test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant. a.u., arbitrary units; EF, ejection fraction; exp., expression; FC, fold change; LVAW, anterior wall of the left ventricle at systole; LVPW, posterior wall of the left ventricle at systole; Rel., relative.
Brca1 promotes Circ-INSR formation during doxorubicin-induced cardiotoxicity
To investigate the upstream regulators of Circ-INSR, we used the RegRNA 2.0 software tool to predict the potential splicing regulatory motifs (Figure 5A). Since, the ALU elements facilitate circRNA formation, we predominantly looked at enhancers, which interact exactly within the circularization elements of Circ-INSR and found Brca1 to be the most promising candidate based on the binding prediction values.
Figure 5.
Circ-INSR is regulated by Brca1. (A) Prediction of the upstream regulator of Circ-INSR using the RegRNA2.0 tool. (B) Relative expression of Brca1 in mouse heart tissue with or without doxorubicin treatment (n = 6 mice per group). (C) Relative expression of Brca1 in HL-1 cells treated with Brca1-siRNA compared with Scramble siRNA (n = 7, from two independent experiments). (D) Relative expression of Circ-INSR and INSR in HL-1 cells treated with Brca1-siRNA compared with Scramble siRNA (n = 7, from two independent experiments). (E) Relative expression of Circ-INSR and INSR in pcDNA-Circ-INSR transfected HL-1 cells co-treated with Brca1-siRNA compared with control transfected with pcDNA-Empty and Scramble siRNA (n = 7, from two independent experiments, one-way analysis of variance performed within Control, pcDNA-Circ-INSR + Scramble siRNA, and pcDNA-Circ-INSR + Brca1-siRNA). All quantitative data are presented as mean ± standard error mean, and an unpaired two-tailed t-test was performed to calculate significance between two groups and one-way analysis of variance with post hoc Tukey test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant. exp., expression; FC, fold change; Rel., relative.
Interestingly, the decreased expression of Circ-INSR in the hearts of doxorubicin-treated mice was paralleled by a down-regulation of Brca1 expression (Figure 5B). The siRNAs designed against Brca1 efficiently blocked Brca1 expression in HL-1 cardiomyocytes (Figure 5C) which led to a significantly reduced expression of Circ-INSR without affecting the host gene INSR (Figure 5D). Co-transfection of a Circ-INSR overexpression plasmid and Brca1 siRNAs into HL-1 cardiomyocytes resulted in significantly lower Circ-INSR levels compared with co-transfection with control siRNAs, demonstrating that Brca1 facilitates Circ-INSR formation (Figure 5E). These results suggest Brca1 as a modulator of Circ-INSR expression which is independent of the host gene.
Circ-INSR physically interacts with single-stranded DNA-binding protein 1 to regulate mitochondrial function in cardiomyocytes
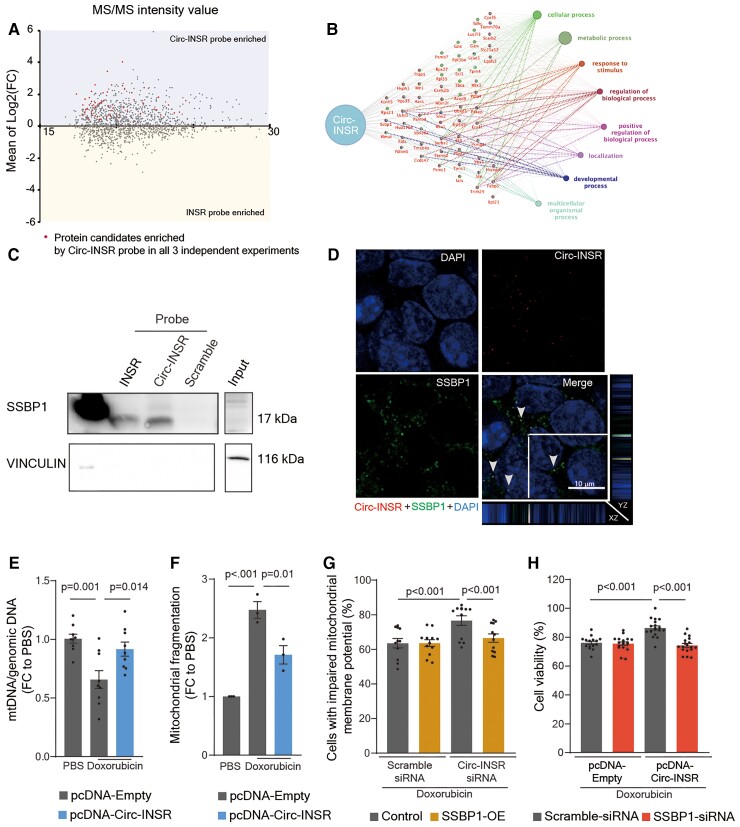
To investigate underlying mechanisms of Circ-INSR-mediated cardioprotection, we aimed to screen for proteins that explicitly interact with Circ-INSR. We designed two different biotin-labelled probes which could hybridize either to the back-splice junction of Circ-INSR or to the linear INSR mRNA (Supplementary material online, Figure S4A). The RT-qPCR experiments validated the specificity and efficiency of the probes (Supplementary material online, Figure S4B). Mass spectrometry analysis of the pulldown lysates identified potential Circ-INSR-bound proteins. Fifty-eight proteins were preferentially enriched by the Circ-INSR probe compared with the linear INSR mRNA probe consistently in all three independent pulldown sets analysed in the mass spectrometry (Figure 6A). These 58 Circ-INSR associated proteins were further used to construct a pathway cluster network for Gene Ontology (GO) using Cluego.33 In line with the RNA sequencing analysis, after Circ-INSR knockdown, the most enriched GO term was ‘metabolic process’ (Figure 6B). The 58 candidates were additionally subjected to in silico prediction for their interaction probability with Circ-INSR by applying the CatRAPID algorithm,34 which yielded single-stranded DNA-binding protein 1 (SSBP1) as the strongest possible binding partner based on fold change and interaction propensity (Supplementary material online, Figure S4C, Table S2). The physical interaction between Circ-INSR and SSBP1 was validated in RNA pulldown experiments (Figure 6C) as well as SSBP1 immunoprecipitation experiments (Supplementary material online, Figure S4D). In addition, immuno-RNA-FISH staining revealed that the Circ-INSR-specific RNA-FISH signals co-localize with SSBP1 immunofluorescence signals in the cytoplasm (Figure 6D, grey arrows) further corroborating a strong direct interaction.
Figure 6.
Circ-INSR regulates metabolism and cell viability via interaction with single-stranded DNA-binding protein 1 in cardiomyocytes. (A) MA plot showing the relationship between intensity value and fold change (log2FC) among the proteins. Only those proteins which were enriched by the Circ-INSR probe in all three individual experiments were labelled with red dots. (B) Interaction network laid out and visualized with ClueGO plugin of Cytoscape. Dots indicate the proteins interacting with Circ-INSR, and the respective colours indicate the interaction types. Each gene ontology pathway is depicted by a specific node colour. The size of the node represents the number of interacting proteins, which belong to the respective gene ontology pathway. (C) Western blot of single-stranded DNA-binding protein 1 levels in HL-1 cells after Circ-INSR and INSR RNA pulldown using biotin-labelled DNA probes. (D) RNA fluorescence in situ hybridization (RNA-FISH) of Circ-INSR (red) and co-staining with single-stranded DNA-binding protein 1 (green) and DAPI (blue) in HL-1 cells (scale bar, 10 μm); Z-stack view of the co-localized single-stranded DNA-binding protein 1 and Circ-INSR molecules are shown around the edge of the merge figure panel. Grey arrows highlight the yellow dots which indicate the co-localization of Circ-INSR and single-stranded DNA-binding protein 1 in the cytoplasm. (E) Mitochondrial DNA (mtDNA) copy number analysis of HL-1 cells treated with pcDNA-Circ-INSR overexpression plasmid and doxorubicin compared with pcDNA-Empty control (0.25 μM 48 h, n = 9 wells per group from triplicates of three independent experiments, one-way analysis of variance was performed). (F) Quantification of MitoTracker staining assay in HL-1 cells treated with pcDNA-Circ-INSR overexpression plasmid and doxorubicin compared with pcDNA-Empty control (0.25 μM 48 h, n = 3 independent experiments, one-way analysis of variance performed within pcDNA-Empty, pcDNA-Empty + Doxorubicin, and pcDNA-Circ-INSR + Doxorubicin). (G) Mitochondrial membrane potential assay in HL-1 cells treated with Circ-INSR-siRNA, single-stranded DNA-binding protein 1-overexpression plasmid under doxorubicin stress (0.25 μM 48 h, n = 11/12/12/12 from three independent experiments, two-way analysis of variance, Sidak’s multiple comparison test was performed). Percentage of cells with impaired mitochondrial membrane potential is plotted on the graphs. (H) WST assay in HL-1 cells treated with pcDNA-Circ-INSR overexpression plasmids, single-stranded DNA-binding protein 1-siRNA under doxorubicin stimulation (0.25 μM 48 h, n = 15/18/18/18 from three independent experiments, two-way analysis of variance, Tukey’s multiple comparison test was performed). All quantitative data are presented as mean ± standard error mean, one-way analysis of variance with post hoc Tukey test was used to calculate significance between ≥3 groups and two-way analysis of variance with Sidak’s or Tukey’s multiple comparison test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant. FC, fold change; Rel., relative; exp., expression; MS/MS, tandem mass spectrometry; OE, overexpression; SSBP1, single-stranded DNA-binding protein.
The SSBP1 is known to regulate mitochondrial DNA (mtDNA) stability35 and replication.36 Based on the mitochondrial phenotype observed in Circ-INSR gain- or loss-of-function experiments, we hypothesized that Circ-INSR may cooperate with SSBP1 resulting in the regulation of mitochondrial function by stabilizing mtDNA. Indeed, the mtDNA content strongly decreased in cardiomyocytes treated with doxorubicin, but was preserved upon Circ-INSR overexpression (Figure 6E). This directly correlated with changes in the mitochondrial morphology as indicated by mitochondrial fragmentation, which was significantly prevented in cardiomyocytes overexpressing Circ-INSR under doxorubicin stress (Figure 6F and Supplementary material online, Figure S4E). To further dissect the functional interaction between SSBP1 and Circ-INSR, we established SSBP1 overexpression and knockdown system in HL-1 cells (Supplementary material online, Figure S4F and G). Then, we applied SSBP1 overexpression and measured the mitochondrial membrane potential as a functional read-out, using doxorubicin to induce impairment of the mitochondrial membrane potential. Circ-INSR knockdown exacerbated the damage to the mitochondrial membrane potential. Strikingly, simultaneous overexpression of SSBP1 blocked this deleterious effect (Figure 6G and Supplementary material online, Figure S4H). Conversely, silencing of SSBP1 nullified the cardioprotective effects of Circ-INSR overexpression under doxorubicin stress, thus confirming a direct role for SSBP1 downstream of Circ-INSR (Figure 6H).
These mechanistic data demonstrate that Circ-INSR interacts and synergizes with SSBP1 to achieve its protective effects in the context of doxorubicin-induced mitochondrial dysfunction and apoptosis in cardiomyocytes.
Specificity and translation of Circ-INSR-mediated cardioprotective effects to human pre-clinical models
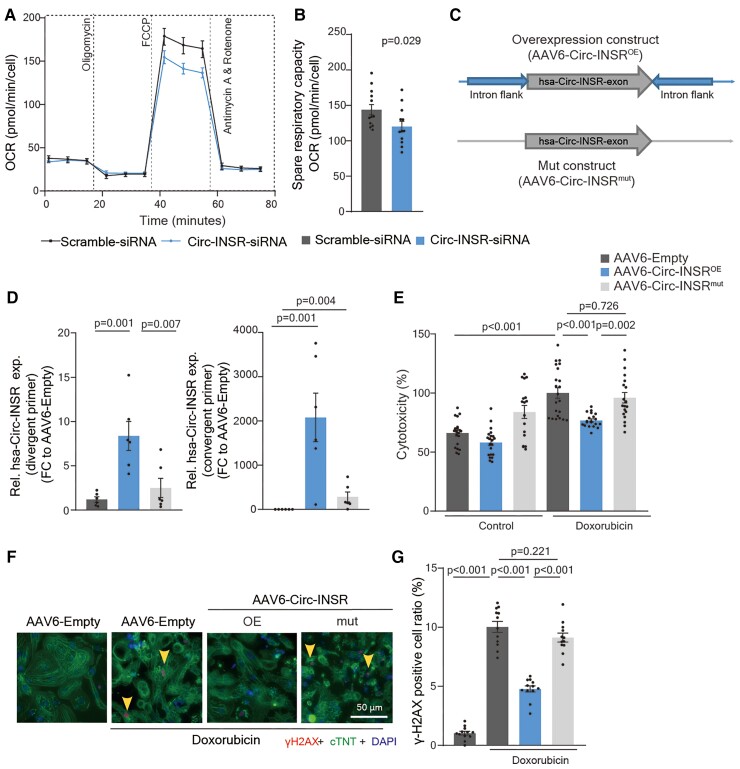
To test the translational value of Circ-INSR therapy, we evaluated Circ-INSR function in human cardiomyocytes. Interestingly, Circ-INSR siRNA2 inhibited the Circ-INSR expression in human-induced pluripotent stem-cell-derived cardiomyocytes without affecting the expression of human INSR. We observed that the Circ-INSR silencing (Supplementary material online, Figure S5A) impaired metabolic activity in human-induced pluripotent stem-cell-derived cardiomyocytes as indicated by significant reduction of oxygen consumption rate and spare respiratory capacity (Figure 7A and B).
Figure 7.
Human Circ-INSR regulates metabolic activity and prevents cell death in human cardiomyocytes. (A and B) Mean oxygen consumption rate at various time points of Mito Stress Assay and spare respiratory capacity of hiPSC-CMs treated with Circ-INSR-siRNA compared with Scramble siRNA (n = 12 wells/group from one experiment). (C) Schematic representation of human Circ-INSR overexpression and mut construct. (D) Circ-INSR expression (using divergent and convergent primer pairs) in hiPSC-CMs after AAV6-Circ-INSROE transduction compared with AAV6-Empty and AAV6-Circ-INSRmut (n = 6 per group, one-way analysis of variance was performed within AAV6-Empty, AAV6-Circ-INSROE and AAV6-Circ-INSRmut). (E) Percentage of cell cytotoxicity (LDH assay) in hiPSC-CMs transduced with AAV6-Circ-INSROE compared with AAV6-Empty and AAV6-Circ-INSRmut in presence or absence of doxorubicin (1 μM 48 h, n = 20/21/17/22/20/19 from two independent experiments, one-way analysis of variance was performed within AAV6-Empty + Doxorubicin, AAV6-Circ-INSROE + Doxorubicin, and AAV6-Circ-INSRmut + Doxorubicin, unpaired two-tailed t-test was performed between AAV6-Empty and AAV6-Empty + Doxorubicin). (F and G) γ-H2AX (red) staining in nuclei (DAPI, blue) of hiPSC-CMs (cTNT, green) transduced with either AAV6-Empty control, AAV6-Circ-INSROE, or AAV6-Circ-INSRmut in the presence or absence of doxorubicin (1 μM 48 h, n = 12 per group, from three independent experiments, scale bar, 50 μm, one-way analysis of variance was performed within AAV6-Empty + Doxorubicin, AAV6-Circ-INSROE + Doxorubicin, and AAV6-Circ-INSRmut + Doxorubicin, unpaired two-tailed t-test was performed between AAV6-Empty and AAV6-Empty + Doxorubicin). Yellow arrows denote DNA damage. Representative images left; quantification right. All quantitative data are presented as mean ± standard error mean, and an unpaired two-tailed t-test was performed to calculate significance between two groups, and one-way analysis of variance with post hoc Tukey test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant. exp., expression; FC, fold change; hiPSC-CM, human-induced pluripotent stem-cell-derived cardiomyocyte; OCR, oxygen consumption rate; Rel., relative.
We next generated a human Circ-INSR overexpression (Circ-INSROE) construct as well as a construct lacking circularization elements (Circ-INSRmut; Figure 7C). We tested the overexpression of human Circ-INSR at various multiplicity of infection (MOI) of AAV6-Circ-INSROE (Supplementary material online, Figure S5B) and demonstrated the protective effects against doxorubicin-induced cardiomyocyte apoptosis at an MOI of 104 and 105 (Supplementary material online, Figure S5C).
Furthermore, AAV6-mediated cardiomyocyte transduction with Circ-INSROE but not Circ-INSRmut led to a robust Circ-INSR up-regulation (Supplementary material online, Figure S5D). Remarkably, AAV-based overexpression of human Circ-INSR significantly attenuated doxorubicin-induced apoptosis in primary rat cardiomyocytes, while the circularization deficient construct did not (Supplementary material online, Figure S5E–G). Similarly, human-induced pluripotent stem-cell-derived cardiomyocytes infected with AAV6-Circ-INSROE but not AAV6-Circ-INSRmut provided protection against doxorubicin-mediated DNA damage indicated by γH2AX analysis (Figure 7D–G).
Collectively, protection from doxorubicin-mediated cardiotoxicity in primary rat and human cardiomyocytes requires the circularized form of Circ-INSR.
In vitro transcribed Circ-INSR protects against doxorubicin-mediated cardiomyocyte death
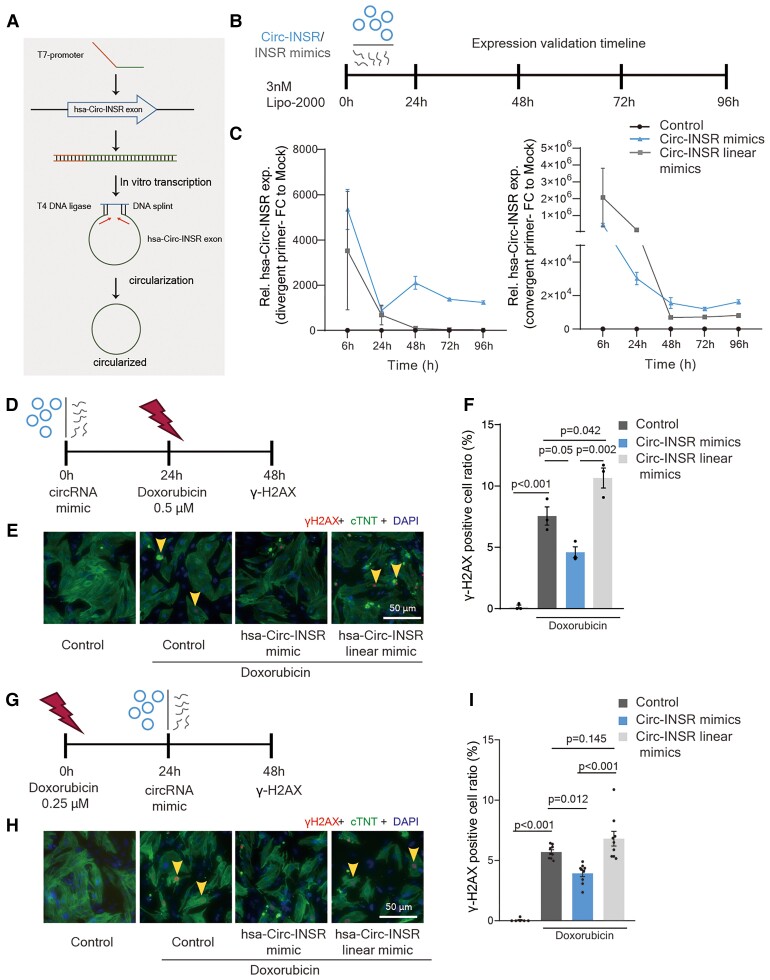
Based on the promising translational data of Circ-INSR, we aimed to generate a potential RNA therapy for clinical application against doxorubicin-induced cardiotoxicity. Hence, we produced human Circ-INSR mimics via in vitro transcription and subsequent circularization via a DNA splint and enzymatic ligation (Figure 8A and Supplementary material online, Figure S6A–C). Circularized Circ-INSR mimics as well as the linear Circ-INSR transcripts were transfected into primary rat cardiomyocytes (Figure 8B). Circ-INSR levels persisted over time indicating stability of in vitro transcribed and circularized Circ-INSR (Figure 8C). To test the efficiency, we first delivered these in vitro produced Circ-INSR mimics to primary rat cardiomyocytes before doxorubicin treatment (Figure 8D). Circ-INSR mimics prevented doxorubicin-mediated DNA damage compared with the control and Circ-INSR linear mimics (Figure 8E and F). Finally, we also tested the therapeutic potential of in vitro transcribed Circ-INSR in primary rat cardiomyocytes post-doxorubicin stress (Figure 8G). Strikingly, in vitro transcribed Circ-INSR demonstrated a strong cardioprotective effect on doxorubicin-induced cardiomyocyte DNA damage (Figure 8H and I).
Figure 8.
In vitro transcribed Circ-INSR mimics mediated therapy in cardiomyocytes against doxorubicin cardiotoxicity. (A) Schematic representation of in vitro production and circularization of Circ-INSR mimics. (B) Schematic depiction of Circ-INSR mimic and Circ-INSR linear mimics transfection to neonatal rat cardiomyocytes via lipo-2000. (C) Circ-INSR expression (using divergent and convergent primer pairs) in neonatal rat cardiomyocytes after Circ-INSR mimics transfection compared with control and Circ-INSR linear mimics (n = 3–4 per group) at different timepoints. (D) Schematic representation of preventive therapy approach using Circ-INSR mimics. (E and F) γ-H2AX (red) staining in nuclei (DAPI, blue) of neonatal rat cardiomyocytes (cTNT, green) transfected with Circ-INSR mimics and Circ-INSR linear mimics compared with control, under doxorubicin treatment. Yellow arrows denote DNA damage. Representative images left; quantification right (0.5 μM 48 h, n = 3 per group, scale bar, 50 μm, one-way analysis of variance was performed within Control + Doxorubicin, Circ-INSR mimics + Doxorubicin, and Circ-INSR linear mimics + Doxorubicin, unpaired two-tailed t-test was performed between Control and Control + Doxorubicin). (G) Schematic representation of therapeutic strategy using Circ-INSR mimics; (H and I) γ-H2AX (red) staining in nuclei (DAPI, blue) of neonatal rat cardiomyocytes (cTNT, green) transfected with Circ-INSR mimics and Circ-INSR linear mimics compared with control, in the presence or absence of doxorubicin. Yellow arrows denote DNA damage. Representative images left; quantification right (0.2 μM 48 h, n = 6–9 per group, from three independent experiments (scale bar, 50 μm), one-way analysis of variance was performed within Control + Doxorubicin, Circ-INSR mimics + Doxorubicin, and Circ-INSR linear mimics + Doxorubicin, unpaired two-tailed t-test was performed between Control and Control + Doxorubicin). All quantitative data are presented as mean ± standard error mean, an unpaired two-tailed t-test was performed to calculate significance between two groups, and one-way analysis of variance with post hoc Tukey test was used to calculate significance between ≥3 groups wherever required. P < 0.05 was considered as statistically significant. exp., expression; FC, fold change; NRCMs, neonatal rat cardiomyocytes; Rel., relative.
In summary, delivery of in vitro transcribed Circ-INSR mimics efficiently protects against doxorubicin-induced cardiotoxicity which presents an exciting complementary alternative for AAV-based delivery.
Discussion
Here we report a significant protective effect of the highly species-conserved Circ-INSR against doxorubicin-mediated cardiotoxicity in rodent and human cardiomyocytes in vitro as well as in a chronic doxorubicin-induced cardiotoxicity mouse model. We identified a physical and functional interaction of Circ-INSR with SSBP1 which plays a role in modulating cardiomyocyte apoptosis and mitochondrial membrane potential. In addition to AAV-mediated Circ-INSR overexpression in vitro and in vivo, we also applied the in vitro transcribed and circularized Circ-INSR mimics. All approaches efficiently prevented doxorubicin-mediated cardiotoxicity highlighting the therapeutic value of Circ-INSR (Structured Graphical abstract).
Apart from doxorubicin, other anthracyclines or anti-cancer drugs such as taxanes are known to induce unexpected dose- and time-dependent cardiotoxicity.37–39 Therefore, we also tested Circ-INSR expression after treatment of cardiomyocytes with different concentrations of other anti-cancer drugs such as epirubicin and paclitaxel (Supplementary material online, Figure S6D and E). Our data suggest that Circ-INSR may be important in other cardiotoxicity models as well, but further detailed investigation is needed in this context.
Brca1 is a well-established tumour suppressor gene.40 Besides its specific role in DNA double-strand break repair,41 it was also identified as an exonic splicing enhancer.42 Loss of Brca1 was shown to weaken the process of double-strand DNA break repair and to activate the p53-mediated apoptosis pathway in cardiomyocytes.43 Our data showed lower levels of Brca1 in doxorubicin-treated heart tissue. Brca1 silencing led to diminished expression of Circ-INSR highlighting a putative role of Brca1 in the generation of circular transcripts from the INSR locus. However, the exact mechanism by which Brca1 regulates the circularization of Circ-INSR transcript is still unknown and requires further mechanistic studies.
Previous evidence suggests that circRNAs play specific roles in a variety of cardiac diseases, although detailed information is scarce for this rather recently discovered type of ncRNAs.17,44 Cdr1as, which contains more than 70 conserved miR-7 target sites, was identified as a miR-7 repressor in the brain and is the most studied circRNA molecule to date.45 Another study revealed that Cdr1as could function as a powerful miR-7a sponge in the mouse heart resulting in the promotion of myocardial infarction.46 To date, the majority of reports on circRNAs suggest such a competing endogenous RNA mechanism. However, in contrast to the 70 miR-7 binding sites present on Cdr1as, most circRNAs often possess only a few complementary-binding sites for their target microRNAs. In this context, Circ-INSR was recently reported to participate in skeletal muscle myoblast proliferation through an intricate mechanism that involves sponging of miR-34a and miR-15b.47 In contrast, for Circ-INSR function in the heart muscle, we do not find evidence for competing endogenous RNA mechanisms. Circ-INSR gain and loss-of function experiments did not regulate miR-34a, miR-15b, and miR-221 expression in HL-1 cells (Supplementary material online, Figure S6F and G). Furthermore, RNA pulldown experiments of Circ-INSR did not reveal co-precipitated miR-34 in HL-1 cells (Supplementary material online, Figure S6H). It is at least debatable whether microRNA sponging is a true physiological event based upon similar stoichiometry between the circRNA and microRNAs.48 Here, we demonstrated that Circ-INSR mediates its cardioprotective function through physical interaction with SSBP1, which is a key protein engaged in the process of mtDNA replication and protection. The SSBP1 can regulate the stability of single-stranded mtDNA following mtDNA synthesis stimulated by the DNA polymerase subunit gamma.49 The SSBP1 is also known to participate in the process of angiotensin II-induced cardiac fibrosis and loss of SSBP1 enhances the expression of fibrosis markers.50 Another investigation reported that SSBP1 was up-regulated in cardiomyocytes with the treatment of Qiliqiangxin, a traditional Chinese medicine showing protective effects in a clinical trial in HF patients.51 We reported here that Circ-INSR regulates mitochondrial functions, such as mitochondrial fragmentation, metabolism, and mtDNA replication in cardiomyocytes under doxorubicin stress. In addition, we are the first to provide direct evidence that SSBP1 binds to Circ-INSR by circRNA pulldown, SSBP1 immunoprecipitation, and RNA-FISH experiments. Inhibition of SSBP1 abrogates the cardioprotection of Circ-INSR overexpression, whereas mitochondrial impairment after Circ-INSR knockdown can be rescued by SSBP1 overexpression, thus suggesting also a functional interaction.
One of the challenges of ncRNA-based therapeutics (especially for long ncRNAs) is the poor conservation across species.52 However, circRNAs are a specific class of ncRNAs which possess a high degree of conservation most likely due to the fact that majority of circRNAs originate from precursor mRNA.17 Indeed, we observed Circ-INSR to exhibit an 85% sequence similarity between human and rodent transcripts. In addition to species-specific Circ-INSR overexpression, we also observed cross-species complementation as indicated by cardioprotection under doxorubicin stress after overexpression of human Circ-INSR in primary rodent cardiomyocytes and vice versa. Of note, there are fundamental differences between adult cardiomyocytes and primary (neonatal) cardiomyocytes or hiPSC-derived cardiomyocytes. As the latter tend to be structurally and functionally more immature, caution is warranted when interpreting results.53 Nevertheless, since the results are paralleled in the adult mouse heart in vivo, these pre-clinical data suggest the excellent translational potential of Circ-INSR in future pre-clinical and clinical development processes.54
An important concern for applying circRNA overexpression is the ‘contamination’ by linear isoforms, which may be produced as by-products during the conventional circRNA overexpression approach. Therefore, we designed a Circ-INSR overexpression plasmid which lacks circularization elements, thus inhibiting the formation of circular products. Interestingly, the linear transcript of Circ-INSR RNA did not demonstrate any therapeutic effects compared with the circularized Circ-INSR in cardiomyocytes indicating that the protective effect of Circ-INSR requires its circular structure.
Moreover, technological advances such as in vitro transcription, including additional steps for the circularization and purification of circRNA transcripts, may improve future production of precise and specific circRNA oligonucleotides.55–58 To further strengthen our findings, we produced in vitro transcribed circRNA mimics and circularized the transcripts via DNA splint and T4 DNA ligase. Our results show that in vitro transcribed Circ-INSR mimics can prevent doxorubicin-mediated cardiomyocyte apoptosis. As in vitro transcribed circRNA mimics can exert their function instantly (when compared with transgene delivery/expression), and even remain stable in cells, our study provides the first proof of concept for researchers to apply in vitro transcribed circRNAs directly as RNA therapeutics targeted to CVDs.
In summary, Circ-INSR rescues doxorubicin-induced cardiac dysfunction and cardiomyocyte apoptosis through interaction with SSBP1, thus maintaining mitochondrial function. Further studies are warranted to develop this novel target and its overexpression strategy into a clinical preventive therapy for patients at high risk for doxorubicin-mediated cardiotoxicity. Altogether, this study reveals Circ-INSR as a promising target for the prevention of anti-cancer drug-induced cardiotoxicity.
Author contributions
D.C.L., S.Ch., and K.X. performed the experiments, analysed the data, and drafted the manuscript. I.R., C.K.H, A.C., S.Cu., D.N., L.R., A.S., M.J., J.L., O.L.G., and U.K. provided technical assistance. J.B. and G.B. provided support for planning and executing in vivo experiment and data analysis. W.L.W.T. and R.F. performed circRNA sequencing. A.V., L.W.V.L., and P.V.D.M. provided clinical samples. C.B. and T.T. developed the concept, designed the study, and revised the manuscript. All authors have read, discussed the results, and approved the manuscript.
Supplementary data
Supplementary data are available at European Heart Journal online.
Supplementary Material
Acknowledgements
The authors thank Anna-Maria Bergmann (from IMTTS) for the help of analysis in the mito-fragmentation assay. The authors thank Karina Zimmer and Dr Heike Janssen-Peters (from IMTTS) for help with the planning and execution of animal experiments. They also acknowledge support by the MHH core facilities for Transcriptomics, Proteomics, and Laser Microscopy.
Contributor Information
Dongchao Lu, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; REBIRTH Center for Translational Regenerative Medicine, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Shambhabi Chatterjee, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; REBIRTH Center for Translational Regenerative Medicine, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Ke Xiao, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; Fraunhofer Institute for Toxicology and Experimental Medicine (ITEM), Nikolai-Fuchs-Straße 1, Hannover 30625, Germany.
Isabelle Riedel, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Cheng-Kai Huang, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Alessia Costa, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; REBIRTH Center for Translational Regenerative Medicine, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Sarah Cushman, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Dimyana Neufeldt, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Laura Rode, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Arne Schmidt, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Malte Juchem, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Julia Leonardy, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Gwen Büchler, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Jonas Blume, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Olivia-Luise Gern, Institute for Experimental Infection Research, TWINCORE, Centre for Experimental and Clinical Infection Research, Feodor-Lynen-Straße 7, Hannover 30625, Germany; Department of Pathology, University of Veterinary Medicine Hannover, Foundation, Bünteweg 2, Hannover 30559, Germany.
Ulrich Kalinke, Institute for Experimental Infection Research, TWINCORE, Centre for Experimental and Clinical Infection Research, Feodor-Lynen-Straße 7, Hannover 30625, Germany; Cluster of Excellence—Resolving Infection Susceptibility (RESIST), Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany.
Wilson Lek Wen Tan, Cardiovascular Disease Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Centre for Translational Medicine, 14 Medical Drive, Level 8, Singapore 117599, Republic of Singapore.
Roger Foo, Cardiovascular Disease Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Centre for Translational Medicine, 14 Medical Drive, Level 8, Singapore 117599, Republic of Singapore.
Aryan Vink, Department of Pathology, University Medical Center Utrecht, Heidelberglaan 100, Utrecht 3584, The Netherlands.
Linda W van Laake, Department of Cardiology and Regenerative Medicine Center, University Medical Centre Utrecht, Heidelberglaan 100, Utrecht 3584, The Netherlands.
Peter van der Meer, Department of Cardiology, University of Groningen, University Medical Center Groningen, Hanzeplein 1, Groningen 9713, The Netherlands.
Christian Bär, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; REBIRTH Center for Translational Regenerative Medicine, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; Fraunhofer Institute for Toxicology and Experimental Medicine (ITEM), Nikolai-Fuchs-Straße 1, Hannover 30625, Germany.
Thomas Thum, Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; REBIRTH Center for Translational Regenerative Medicine, Hannover Medical School, Carl-Neuberg-Str. 1, Hannover 30625, Germany; Fraunhofer Institute for Toxicology and Experimental Medicine (ITEM), Nikolai-Fuchs-Straße 1, Hannover 30625, Germany.
Funding
This work was supported through funding from the German Research Foundation, DFG [SFB/Transregio TRR267, to C.B. and T.T.] and from the Niedersächsische Ministerium für Wissenschaft und Kultur, MWK (REBIRTH Anchor project to T.T.) and German Research Foundation, DFG [398066876/GRK 2485/1 (VIPER) to U.K.]. D.C.L. acknowledges the China Scholarship Council for the funding of his PhD position.
Data availability
The circRNA profile generated from human healthy and heart failure samples and the mass spectrometric data produced by Circ-INSR pull down will be made available upon reasonable request. The RNA sequencing profiling data (GEO: GSE199081) are available in the Gene Expression Omnibus.
References
- 1. Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. . Heart disease and stroke statistics-2019 update: a report from the American heart association. Circulation 2019;139:e56–e528. [DOI] [PubMed] [Google Scholar]
- 2. Wang K, Long B, Liu F, Wang J-X, Liu C-Y, Zhao B, et al. . A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602–2611. [DOI] [PubMed] [Google Scholar]
- 3. Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S, et al. . Circular RNAs in heart failure. Eur J Heart Fail 2017;19:701–709. [DOI] [PubMed] [Google Scholar]
- 4. Roca-Alonso L, Pellegrino L, Castellano L, Stebbing J. Breast cancer treatment and adverse cardiac events: what are the molecular mechanisms? Cardiology 2012;122:253–259. [DOI] [PubMed] [Google Scholar]
- 5. Tocchetti CG, Ameri P, de Boer RA, D’Alessandra Y, Russo M, Sorriento D, et al. . Cardiac dysfunction in cancer patients: beyond direct cardiomyocyte damage of anticancer drugs: novel cardio-oncology insights from the joint 2019 meeting of the ESC working groups of myocardial function and cellular biology of the heart. Cardiovasc Res 2020;116:1820–1834. [DOI] [PubMed] [Google Scholar]
- 6. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 7. Lyon AR, Dent S, Stanway S, Earl H, Brezden-Masley C, Cohen-Solal A, et al. . Baseline cardiovascular risk assessment in cancer patients scheduled to receive cardiotoxic cancer therapies: a position statement and new risk assessment tools from the Cardio-oncology Study Group of the Heart Failure Association of the European Society of Cardiology in collaboration with the International Cardio-oncology Society. Eur J Heart Fail 2020;22:1945–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. N Engl J Med 1998;339:900–905. [DOI] [PubMed] [Google Scholar]
- 9. Swain SM, Whaley FS, Ewer MS. Congestive heart failure in patients treated with doxorubicin: a retrospective analysis of three trials. Cancer 2003;97:2869–2879. [DOI] [PubMed] [Google Scholar]
- 10. Von Hoff DD, Layard MW, Basa P, Davis HL Jr, Von Hoff AL, Rozencweig M, et al. . Risk factors for doxorubicin-induced congestive heart failure. Ann Intern Med 1979;91:710–717.. [DOI] [PubMed] [Google Scholar]
- 11. Chatterjee S, Baer C, Thum T. Linc-ing the noncoding genome to heart function: beating hypertrophy. Trends Mol Med 2017;23:577–579. [DOI] [PubMed] [Google Scholar]
- 12. Viereck J, Buhrke A, Foinquinos A, Chatterjee S, Kleeberger JA, Xiao K, et al. . Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur Heart J 2020;41:3462–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, et al. . Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J 2021;42:178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foinquinos A, Batkai S, Genschel C, Viereck J, Rump S, Gyongyosi M, et al. . Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat Commun 2020;11:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baer C, Chatterjee S, Falcao Pires I, Rodrigues P, Sluijter JPG, Boon RA, et al. . Non-coding RNAs: update on mechanisms and therapeutic targets from the ESC working groups of myocardial function and cellular biology of the heart. Cardiovasc Res 2020;116:1805–1819. [DOI] [PubMed] [Google Scholar]
- 16. Batkai S, Genschel C, Viereck J, Rump S, Baer C, Borchert T, et al. . CDR132L improves systolic and diastolic function in a large animal model of chronic heart failure. Eur Heart J 2021;42:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santer L, Baer C, Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol Ther 2019;27:1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297–1325. [DOI] [PubMed] [Google Scholar]
- 19. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. . Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015;58:870–885. [DOI] [PubMed] [Google Scholar]
- 22. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. . Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014;9:1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. . A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018;37:5829–5842. [DOI] [PubMed] [Google Scholar]
- 25. Schneider T, Bindereif A. Circular RNAs: coding or noncoding? Cell Res 2017;27:724–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo CA, Guo S. Insulin receptor substrate signaling controls cardiac energy metabolism and heart failure. J Endocrinol 2017;233:R131–R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thum T, Borlak J. Gene expression in distinct regions of the heart. Lancet 2000;355:979–983. [DOI] [PubMed] [Google Scholar]
- 28. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. . Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating wnt/beta-catenin signaling under fully defined conditions. Nat Protoc 2013;8:162–175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chatterjee S, Hofer T, Costa A, Lu D, Batkai S, Gupta SK, et al. . Telomerase therapy attenuates cardiotoxic effects of doxorubicin. Mol Ther 2021;29:1395–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abe N, Kodama A, Abe H. Preparation of circular RNA in vitro. Methods Mol Biol 2018;1724:181–192. [DOI] [PubMed] [Google Scholar]
- 31. Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, et al. . The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012;3:1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gupta SK, Garg A, Baer C, Chatterjee S, Foinquinos A, Milting H, et al. . Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res 2018;122:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. . ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. CatRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics 2013;29:2928–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaur P, Longley MJ, Pan H, Wang H, Copeland WC. Single-molecule DREEM imaging reveals DNA wrapping around human mitochondrial single-stranded DNA binding protein. Nucleic Acids Res 2018;46:11287–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morin JA, Cerron F, Jarillo J, Beltran-Heredia E, Ciesielski GL, Arias-Gonzalez JR, et al. . DNA synthesis determines the binding mode of the human mitochondrial single-stranded DNA-binding protein. Nucleic Acids Res 2017;45:7237–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferdinandy P, Baczko I, Bencsik P, Giricz Z, Gorbe A, Pacher P, et al. . Definition of hidden drug cardiotoxicity: paradigm change in cardiac safety testing and its clinical implications. Eur Heart J 2019;40:1771–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cappetta D, Rossi F, Piegari E, Quaini F, Berrino L, Urbanek K, et al. . Doxorubicin targets multiple players: a new view of an old problem. Pharmacol Res 2018;127:4–14. [DOI] [PubMed] [Google Scholar]
- 39. Florescu M, Cinteza M, Vinereanu D. Chemotherapy-induced cardiotoxicity. Maedica (Bucur) 2013; 8:59–67. [PMC free article] [PubMed] [Google Scholar]
- 40. Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, et al. . Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet 1999;22:37–43. [DOI] [PubMed] [Google Scholar]
- 41. Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer 2007;7:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raponi M, Kralovicova J, Copson E, Divina P, Eccles D, Johnson P, et al. . Prediction of single-nucleotide substitutions that result in exon skipping: identification of a splicing silencer in BRCA1 exon 6. Hum Mutat 2011;32:436–444. [DOI] [PubMed] [Google Scholar]
- 43. Shukla PC, Singh KK, Quan A, Al-Omran M, Teoh H, Lovren F, et al. . BRCA1 is an essential regulator of heart function and survival following myocardial infarction. Nat Commun 2011;2:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol 2019;16:503–514. [DOI] [PubMed] [Google Scholar]
- 45. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. . Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- 46. Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. . The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 2016;11:e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shen X, Zhang X, Ru W, Huang Y, Lan X, Lei C, et al. . CircINSR promotes proliferation and reduces apoptosis of embryonic myoblasts by sponging miR-34a. Mol Ther Nucleic Acids 2020;19:986–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 2014;54:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miralles Fuste J, Shi Y, Wanrooij S, Zhu X, Jemt E, Persson O, et al. . In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet 2014;10:e1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tian H-P, Sun YH, He L, Yi YF, Gao X, Xu DL. Single-stranded DNA-binding protein 1 abrogates cardiac fibroblast proliferation and collagen expression induced by angiotensin II. Int Heart J 2018;59:1398–1408. [DOI] [PubMed] [Google Scholar]
- 51. Lin S, Wu X, Tao L, Bei Y, Zhang H, Zhou Y, et al. . The metabolic effects of traditional Chinese medication qiliqiangxin on H9C2 cardiomyocytes. Cell Physiol Biochem 2015;37:2246–2256. [DOI] [PubMed] [Google Scholar]
- 52. Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta 2014;1840:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Onodi Z, Visnovitz T, Kiss B, Hambalko S, Koncz A, Agg B, et al. . Systematic transcriptomic and phenotypic characterization of human and murine cardiac myocyte cell lines and primary cardiomyocytes reveals serious limitations and low resemblances to adult cardiac phenotype. J Mol Cell Cardiol 2021;165:19–30. [DOI] [PubMed] [Google Scholar]
- 54. Huang CK, Kafert-Kasting S, Thum T. Preclinical and clinical development of noncoding RNA therapeutics for cardiovascular disease. Circ Res 2020;126:663–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petkovic S, Muller S. RNA Circularization strategies in vivo and in vitro. Nucleic Acids Res 2015;43:2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beaudry D, Perreault JP. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res 1995;23:3064–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 2006;34:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kariko K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res 2011;39:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The circRNA profile generated from human healthy and heart failure samples and the mass spectrometric data produced by Circ-INSR pull down will be made available upon reasonable request. The RNA sequencing profiling data (GEO: GSE199081) are available in the Gene Expression Omnibus.