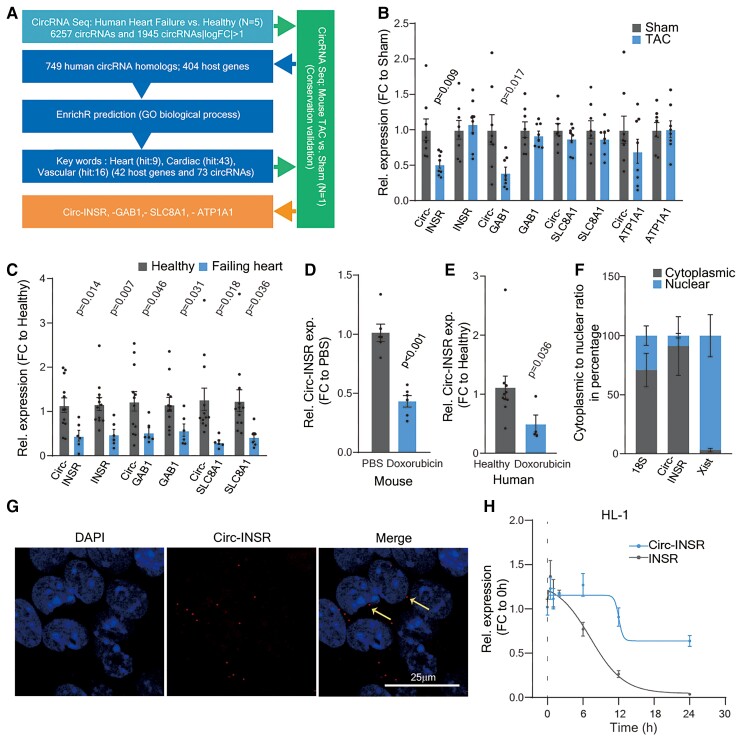
Figure 1.
Identification of Circ-INSR from failing hearts and validation in various heart failure models. (A) Pipeline to select candidates from RNA-Seq data. (B) Relative expression of circRNA and their linear host genes (Circ-INSR, INSR, Circ-GAB1, GAB1, Circ-SLC8A1, SLC8A1, Circ-ATP1A1, and ATP1A1) in TAC model at 13 weeks (n = 8 mice/group). (C) Relative expression of circRNA and their linear host genes (Circ-INSR, INSR, Circ-GAB1, GAB1, Circ-SLC8A1, and SLC8A1) in failing heart (n = 6 patients) compared with healthy (n = 12), expression of Circ-ATP1A1 was not detectable in the human samples. (D) Relative expression of Circ-INSR in mouse hearts treated with doxorubicin (n = 6 per group). (E) Relative expression of Circ-INSR in male patient heart with doxorubicin-induced heart failure (n = 4) compared with healthy (n = 10; Mann–Whitney U test was performed). (F) Circ-INSR expression in neonatal rat cardiomyocytes represented after normalization to the respective subcellular fraction (18S applied for normalization of cytoplasmic fraction, Xist applied for normalization of nuclear fraction, n = 6 per group from duplicates of three independent experiments). (G) RNA fluorescence in situ hybridization (RNA-FISH) of Circ-INSR in HL-1 cells (scale bar, 25 μm). Thin yellow arrows indicate cytoplasmic localization of the Circ-INSR as observed in RNA-FISH experiment. (H) Relative expression of Circ-INSR and INSR in HL-1 cells treated with Actinomycin D (n = 6 per timepoint). All quantitative data are presented as mean ± standard error mean, and an unpaired two-tailed t-test or Mann–Whitney U test was performed to calculate significance between two groups. P < 0.05 was considered as statistically significant. exp., expression; FC, fold change; GO, gene ontology; NRCM, neonatal rat cardiomyocyte; TAC, transverse aortic constriction; Rel., relative.