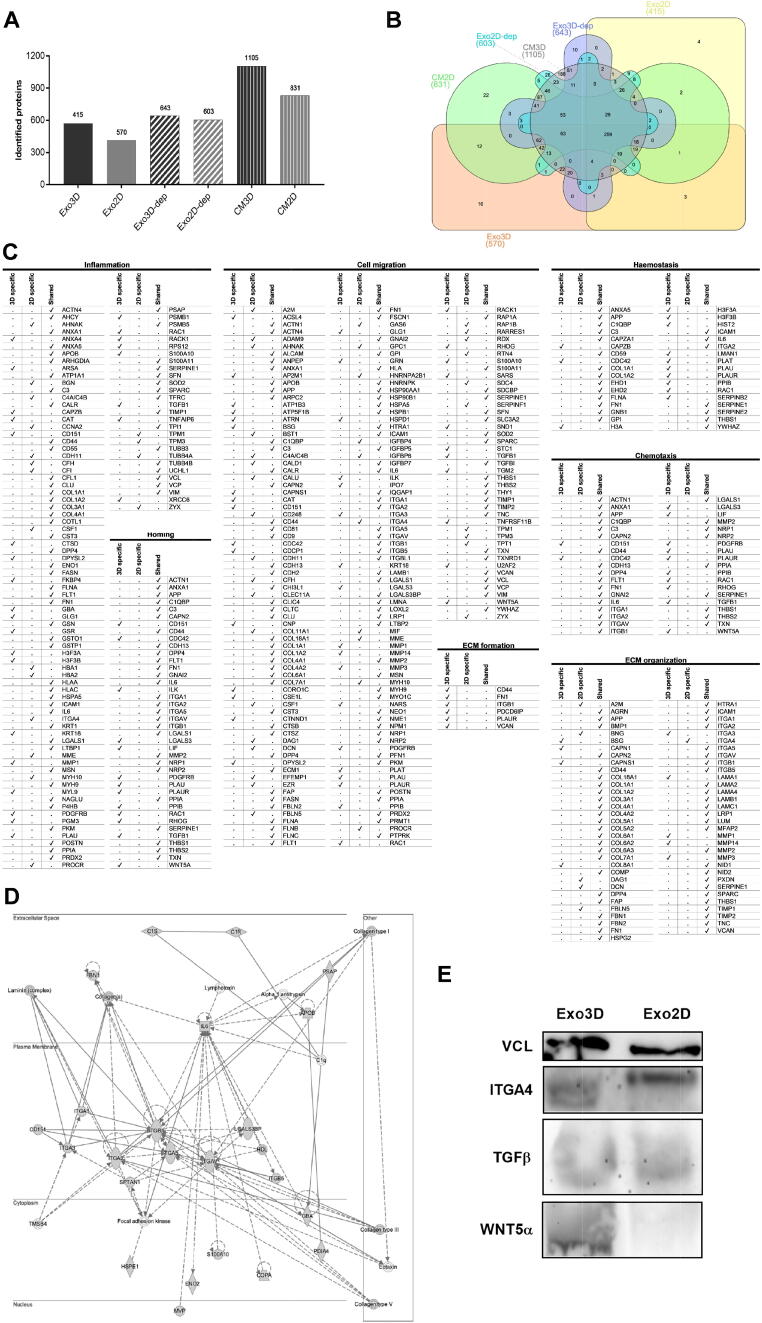
Fig. 2.
Proteomic profiling of MSC-derived secretome and exosomes reveals the presence of proteins related to key biological processes for wound healing. (A) Total protein numbers identified in CM3D, CM2D, Exo3D, Exo2D, Exo3D-dep and Exo2D-dep; (B) Venn diagram shows the unique and shared protein numbers among CM3D, CM2D, Exo3D, Exo2D, Exo3D-dep and Exo2D-dep; (C) IPA Disease&Function analysis for pairwise comparison of Exo3D and Exo2D regarding crucial wound healing events; (D) IPA gene–gene interaction network showing the protein–protein interactions of TGF-β with the Exo3D content regarding the functions of cell morphology, cell-to-cell signalling and interaction, and tissue development; (E) Confirmation of selected proteomic data by western blot analysis of VCL, TGFβ, ITGA4 and WNT5A. Abbreviations: CM3D - whole secretome collected from UC-MSC 3D cultures; CM2D - whole secretome collected from UC-MSC 2D cultures; Exo3D - exosomes derived from UC-MSC cultured in 3D conditions; Exo2D - exosomes derived from UC-MSC cultured in 2D conditions; Exo3D-dep – CM3D depleted of exosomes; Exo2D-dep – CM2D depleted of exosomes.