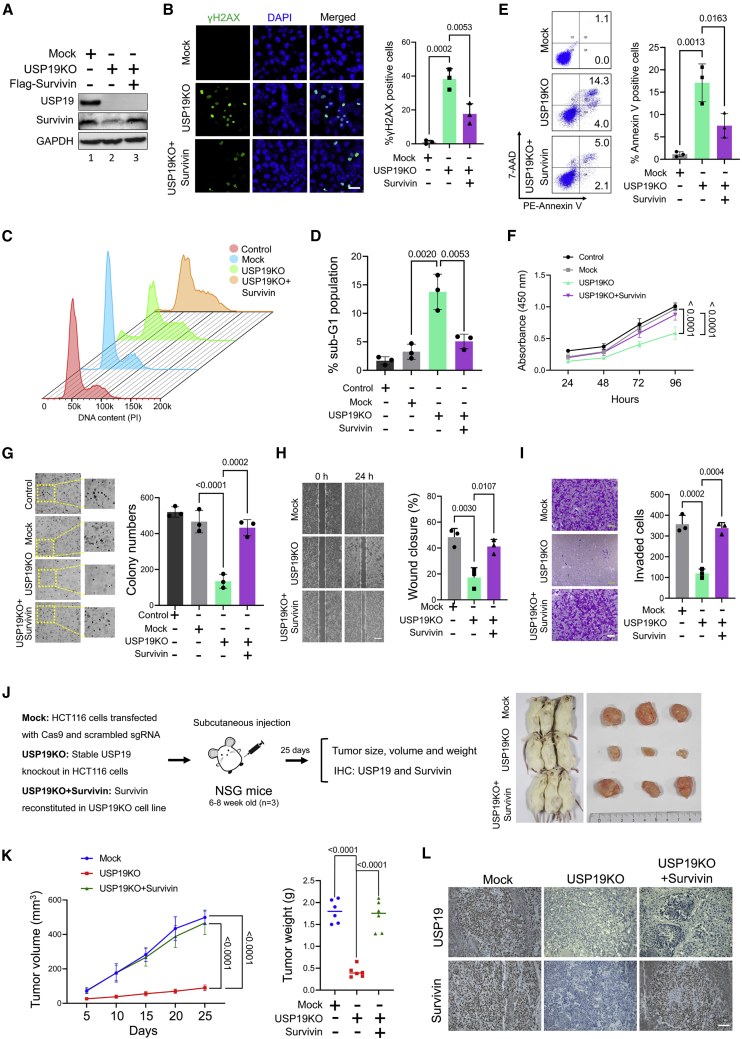
Figure 7.
USP19KO inhibits tumor progression in vitro and in vivo
Mock, USP19KO, and USP19KO cells reconstituted with survivin plasmid in HCT116 cells were used to perform the following experiments. (A) Western blot analysis was performed using USP19- and survivin-specific antibodies. (B) Immunofluorescence analysis was used to measure γH2AX foci formation. Green, γH2AX; blue, nucleus stained by DAPI. Scale bar, 25 μm. The right panel depicts the percentage of γH2AX+ cells. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (C and D) Flow cytometry was performed with PI staining to measure the DNA content and is represented graphically. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (E) Flow cytometry was performed to analyze annexin-V and 7-AAD+ cells and graphically represented. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (F) Cell viability assay was performed using CCK-8 reagent and represented as the absorbance at 450 nm. Data are presented as the means and standard deviations of 3 independent experiments. Two-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (G) Colony formation was measured after 14 days. The colony numbers were quantified and are presented graphically (right panel). Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (H) An in vitro scratch assay was performed to assess the migration potential of the groups mentioned. Scale bar, 100 μm. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (I) Transwell cell invasion assay was performed with the groups mentioned. Scale bar, 200 μm. Invaded cells were quantified using ImageJ software and represented graphically. Data are presented as the means and standard deviations of 3 independent experiments. One-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (J) Xenografts were generated by subcutaneously injecting the mentioned cell groups into the right flank of NSG mice (n = 3). Tumor volumes were recorded and stored for IHC experiments. The right panel shows the tumors excised from the mice after the experiment. (K) Tumor volume and weight were calculated and are presented graphically. Data are presented as the means and standard deviations of 6 biological replicates (n = 6). One-way or 2-way ANOVA followed by Tukey’s post hoc test was used with the indicated p values. (L) Xenograft tumors were sectioned and paraffin embedded. Immunohistochemical analysis was performed with the antibodies indicated. Scale bar, 50 μm.