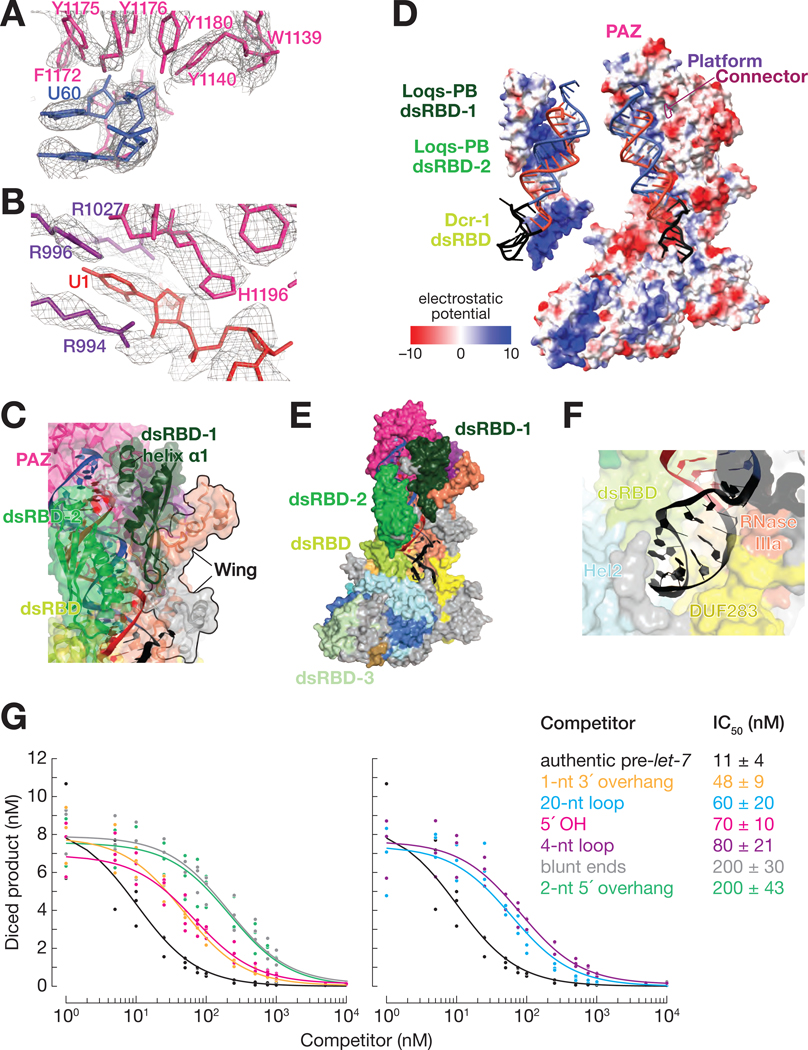
Figure 3. The Dcr-1•Loqs-PB Heterodimer Recognizes All Four Structural Features of Pre-miRNAs.
(A and B) Cryo-EM density (mesh) of the PAZ domain pocket that binds the 3′ overhang of the pre-miRNA (A) and the binding pocket formed by the PAZ and platform domains that binds the 5′ terminal nucleotide (B). Ca2+ ions are shown as gold spheres.
(C) Atomic model of Dcr-1•Loqs-PB binding the stem of the pre-miRNA.
(D) Electrostatic surface view of Dcr-1•Loqs-PB; the RNA is bound in a positively charged channel. Blue, positive charge; red, negative charge.
(E) Atomic model of Dcr-1•Loqs-PB binding the single-stranded loop of the pre-miRNA.
(F) Structure of Dcr-1•Loqs-PB•pre-miRNA complex showing multiple domains of Dcr-1 and Loqs-PB enveloping the pre-miRNA. Protein in surface view; RNA in cartoon view All panels correspond to structure IIa.
(G) Authentic, 5′ 32P-radiolabled pre-let-7 (25 nM) was incubated with Dcr-1•Loqs-PB (5 nM) for 15 min in presence of increasing concentrations of hairpin RNAs, then analyzed by denaturing gel electrophoresis and quantified. Product formed as a function of competitor concentrations is shown for three independent trials. Data were fitted to a 3-parameter inhibition model to obtain the IC50 values. Data are mean ± SD for three independent trials.
See also Figure S6.