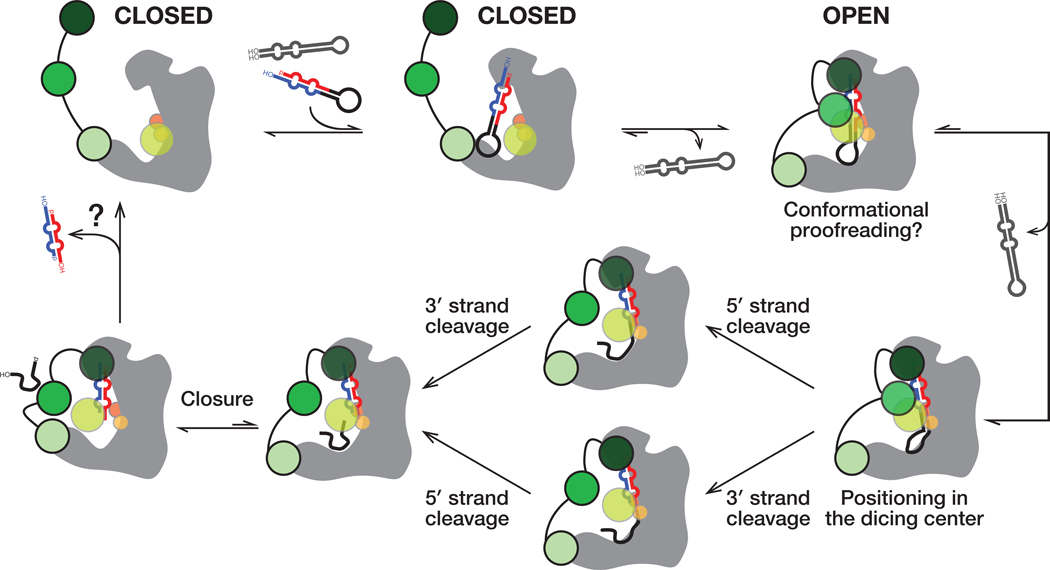
Figure 7. A Model for pre-miRNA processing by the Dcr-1•Loqs-PB heterodimer.
Without pre-miRNA, the closed conformation of the Dcr-1•Loqs-PB complex predominates. Specific interactions between an authentic pre-miRNA and the heterodimer stabilize an open, catalytically competent state. RNA hairpins lacking the pre-miRNA structural features bind weakly and dissociate without cleavage, allowing the heterodimer to return to its closed state. The Dcr-1•Loqs-PB complex envelopes the pre-miRNA. Contacts between RNA and protein residues stabilize a distorted form of pre-miRNA, docking the RNA substrate within the Dcr-1 catalytic centers. Cleavage of the 5′ and 3′ arms of pre-miRNA generates a mature miRNA/miRNA* duplex, allowing the residual loop to depart: partial closure of Dcr-1 is coupled with release of the loop. The miRNA/miRNA* duplex remains anchored to the PAZ and platform domains of Dcr-1 and is released last.