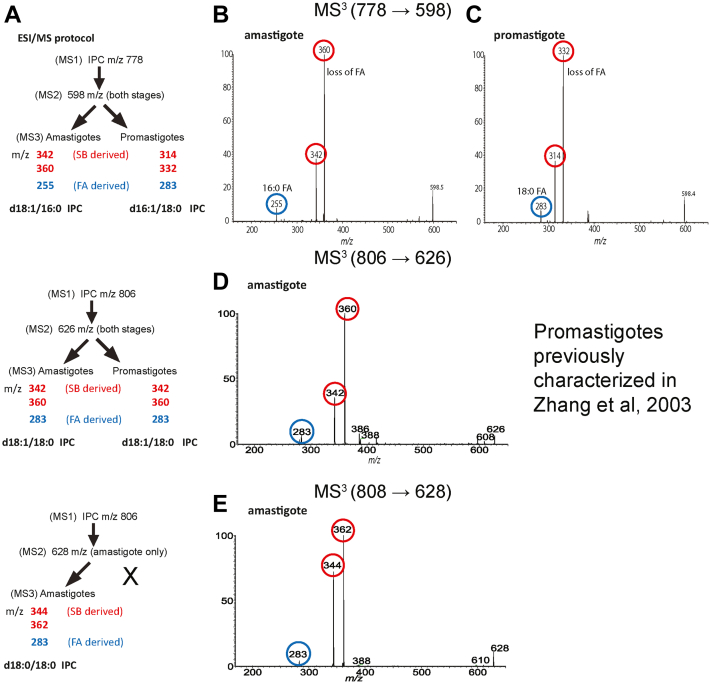
Figure 9.
Characterization of IPC ceramide anchors.A, summary of ESI/MS fragmentation results and deduced ceramide anchor structures. B and C, MS3 (m/z 778.6 → m/z 598) analysis of the IPC ceramide moiety of m/z 598 from amastigotes (B) or promastigotes (C). Unlike promastigote d16:1/18:0-IPC, the deduced amastigote IPC anchor is the same as that of mammalian d18:1/16:0-SM. D, MS3 (m/z 806.6 → m/z 626) analysis of the IPC ceramide moiety of m/z 626 from amastigotes defined the d18:1/18:0-IPC structure, identical to that previously reported for promastigotes (32). E, MS3 (m/z 808.6 → m/z 628) analysis of the amastigote IPC ceramide moiety of m/z 628 (not seen in promastigotes) defined the d18:0/18:0 IPC structure. Fragment ions derived from the fatty acid moiety are highlighted or circled in blue, while SB-derived moieties (acid or ketene as described in the text) are highlighted or circled in red. MS, mass spectrometry; IPC, inositol phosphorylceramide; SB, sphingoid base; SM, sphingomyelin.